


4.7.2. Analysis of variance for a simple lattice design
The steps involved in the analysis of variance when the basic design of simple lattice is repeated only once, are indicated below along with the checks on the computations where they are found important. The material drawn for this illustration is from an experiment conducted at Vallakkadavu, in Kerala involving 25 clones of Eucalyptus grandis.
Table 4.32 below shows the actual field layout showing positions of the blocks and allocation of treatments within each block under randomized condition. The value in the top left corner of each cell is the clone number and the value in bottom right corner shows the mean height of trees in the plot, one year after planting. Unlike in the case of complete block designs, the analysis of variance for incomplete complete block designs involves adjustments on sums of squares for treatments and blocks because of the incomplete structure of the blocks.
Table 4.32. Layout plan for 5 x 5 double lattice showing the height growth (cm) of Eucalyptus grandis clones.
Replication - I
|
Block No. 5 |
25 |
24 |
21 |
23 |
22 |
|
96.40 |
107.90 |
119.30 |
134.30 |
129.20 |
|
|
Block No. 4 |
20 |
19 |
18 |
17 |
16 |
|
148.00 |
99.20 |
101.40 |
98.00 |
106.70 |
|
|
Block No. 1 |
5 |
4 |
1 |
3 |
2 |
|
158.00 |
122.50 |
136.70 |
123.60 |
113.50 |
|
|
Block No. 3 |
13 |
14 |
15 |
12 |
11 |
|
126.80 |
101.60 |
111.70 |
117.30 |
108.20 |
|
|
Block No. 2 |
6 |
9 |
7 |
10 |
8 |
|
126.80 |
127.00 |
119.10 |
90.90 |
130.40 |
Replication - II
|
Block No. 6 |
16 |
6 |
1 |
21 |
11 |
|
169.60 |
157.90 |
124.10 |
134.50 |
112.10 |
|
|
Block No. 9 |
19 |
4 |
9 |
14 |
24 |
|
110.30 |
153.40 |
87.10 |
95.30 |
120.50 |
|
|
Block No. 7 |
7 |
2 |
17 |
22 |
12 |
|
125.60 |
151.10 |
115.90 |
168.40 |
93.30 |
|
|
Block No. 10 |
5 |
20 |
25 |
10 |
15 |
|
126.00 |
106.80 |
137.60 |
132.90 |
117.30 |
|
|
Block No. 8 |
23 |
3 |
8 |
18 |
13 |
|
133.10 |
142.70 |
115.80 |
128.90 |
115.80 |
Step 1. Arrange the blocks within each group (X and Y groups) and treatments within each block systematically along with the observations as in Table 4.33.
Table 4.33. Systematic arrangement of blocks and the treatments within the blocks of Table 4.32.
Replication - I (X-group)
|
Block No. 1 |
1 |
2 |
3 |
4 |
5 |
|
136.70 |
113.50 |
123.60 |
122.50 |
158.00 |
|
|
Block No. 2 |
6 |
7 |
8 |
9 |
10 |
|
126.80 |
119.10 |
130.40 |
127.00 |
90.90 |
|
|
Block No. 3 |
11 |
12 |
13 |
14 |
15 |
|
108.20 |
117.30 |
126.80 |
101.60 |
111.70 |
|
|
Block No. 4 |
16 |
17 |
18 |
19 |
20 |
|
106.70 |
98.00 |
101.40 |
99.20 |
148.00 |
|
|
Block No. 5 |
21 |
22 |
23 |
24 |
25 |
|
119.30 |
129.20 |
134.30 |
107.90 |
96.40 |
Replication - II (Y-group)
|
Block No. 6 |
1 |
6 |
11 |
16 |
21 |
|
124.10 |
157.90 |
112.10 |
169.60 |
134.50 |
|
|
Block No. 7 |
2 |
7 |
12 |
17 |
22 |
|
151.10 |
125.60 |
93.30 |
115.90 |
168.40 |
|
|
Block No. 8 |
3 |
8 |
13 |
18 |
23 |
|
142.70 |
115.80 |
115.80 |
128.90 |
133.10 |
|
|
Block No. 9 |
4 |
9 |
14 |
19 |
24 |
|
153.40 |
87.10 |
95.30 |
110.30 |
120.50 |
|
|
Block No. 10 |
5 |
10 |
15 |
20 |
25 |
|
126.00 |
132.90 |
117.30 |
106.80 |
137.60 |
Step 2. Set up a table of treatment totals by summing up the yields for each clone from both replications. This is shown in Table 4.34. These totals are not adjusted for any block effects.
Table 4.34. Treatment (clone) totals
|
1 |
2 |
3 |
4 |
5 |
|
260.80 |
264.60 |
266.30 |
275.90 |
284.00 |
|
6 |
7 |
8 |
9 |
10 |
|
284.70 |
244.70 |
246.20 |
214.10 |
223.80 |
|
11 |
12 |
13 |
14 |
15 |
|
220.30 |
210.60 |
242.60 |
196.90 |
229.00 |
|
1 6 |
17 |
18 |
19 |
20 |
|
276.30 |
213.90 |
230.30 |
209.50 |
254.80 |
|
21 |
22 |
23 |
24 |
25 |
|
253.80 |
297.60 |
267.40 |
228.40 |
234.00 |
Step 3.Compute the block totals B1, B2, …, B10 for all the blocks by summing the observations occurring in each block. For example, the block total B1 for the first block is given by
B1 = 136.70+113.50+123.60+122.50+158.00 = 654.30
Compute the total for each replication by summing the block totals within each replication. For Replication I,
R1 = B1 + B2 + B3+ B4+ B5 (4.48)
= 654.30 + 594.20 + 565.60 + 553.30 + 587.10
= 2954.50
Compute the grand total as G = R1 + R2 (4.49)
= 2954.50 + 3176.00
= 6130.50
Step 4. Construct an outline for the ANOVA table of simple lattice design.
Table 4.35. Schematic representation of ANOVA table of simple lattice design
|
Source of variation |
Degrees of freedom (df) |
Sum of squares (SS) |
Mean square
|
Computed F |
|
Replication |
r - 1 |
SSR |
MSR |
|
|
Treatment (unadj.) |
k2 - 1 |
SST (unadj.) |
MST (unadj.) |
|
|
Blocks within replication (adj.) |
r(k-1) |
SSB (adj.) |
MSB (adj.) |
|
|
Intra-block error |
(k-1)(rk-k-1) |
SSE |
MSE |
|
|
Total |
rk2 - 1 |
SSTO |
Step 5.Obtain the total sum of squares, replication sum of squares and unadjusted treatment sum of squares. For this, first compute the correction factor (C.F.).
C. F. = ![]() (4.50)
(4.50)
where n = rk2
r = Number of replications
k2 = Number of treatments
k = Number of plots in a block
C. F. = ![]() = 751660.61
= 751660.61
For total sum of squares, find the sum of squares of all the observations in the experiment and subtract the correction factor.
SSTO = ![]() (4.51)
(4.51)
= { (136.70)2 + (113.50)2 +……..+ (137.60)2 } - C. F.
= 770626.43 - 751660.61 = 18965.83
Compute the replication sum of squares as
SSR = ![]() (4.52)
(4.52)
= ![]() 751660.61
751660.61
= 752641.85 - 751660.61 = 981.245
Compute unadjusted treatment sum of squares as
SST (unadj.) = 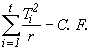 (4.53)
(4.53)
= ![]()
= 760747.90 - 751660.61 = 9087.29
Step 6.Compute for each block, in Replication 1 (X-group) an adjusted block total Cb subtracting each block total in Replication I, from the corresponding column total in Replication II (Y-group) containing the same set of varieties as in blocks of Replication I as shown in Table 4.36. Similarly, compute for each block total in Replication II, an adjusted block total by subtracting each block total in Replication II, from corresponding column total in Replication I (X-group) containing the same set of varieties as in blocks of Replication II as shown in Table 4.37. Obtain the total of Cb values for each replication and check if they add up to zero.
Total of Cb values for Replication I = U1 = 221.50
Total of Cb values for Replication II = U2 = -221.50
This check ensures the arithmetical accuracy of the calculation in the previous steps.
Table 4.36. Computation of Cb values for blocks in Replication I
|
Block |
Replication II Column total |
Replication I Block total |
Cb - value |
|
|
1 |
697.30 |
654.30 |
43.00 |
(C1) |
|
2 |
619.30 |
594.20 |
25.10 |
(C2) |
|
3 |
533.80 |
565.60 |
-31.80 |
(C3) |
|
4 |
631.50 |
553.30 |
78.20 |
(C4) |
|
5 |
694.10 |
587.10 |
107.00 |
(C5) |
|
Total |
3176.00 |
2954.50 |
221.50 |
( |
Table 4.37. Computation of Cb values for blocks in Replication II
|
Block |
Replication I Column total |
Replication II Block total |
Cb- value |
|
|
6 |
597.70 |
698.20 |
-100.50 |
(C6) |
|
7 |
577.10 |
654.30 |
-77.20 |
(C7) |
|
8 |
616.50 |
636.30 |
-19.80 |
(C8) |
|
9 |
558.20 |
566.60 |
-8.40 |
(C9) |
|
10 |
605.00 |
620.60 |
-15.60 |
(C10) |
|
Total |
2954.50 |
3176.00 |
-221.50 |
( |
The adjusted block sum of squares is then given by :
SSB (adj.) = 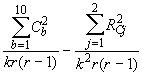 (4.54)
(4.54)
where r = Number of replications,
k = Number of treatments per block.
SSB (adj.) = 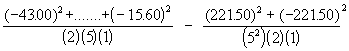
= 3782.05 - 1962.49 = 1819.56
Finally, the error sum of squares is obtained by subtraction.
SSE = SSTO - SSR - SST (unadj.) - SSB (adj.) (4.55)
= 18965.83 - 981.24 - 9087.29 - 1819.56
= 7077.73
Note that the sum of squares due to error (SSE) computed here indicates that part of the variation (in the response variable) between plots within each block caused by uncontrolled extraneous factors and is generally known as the intra-block error. The adjusted block sum of squares is called inter block error.
Step 7.After obtaining the different sums of squares, enter all the values in the ANOVA table as shown as Table 4.38. The mean squares are obtained by dividing the sum of squares by degrees of freedom as usual.
Table 4.38. ANOVA table of simple lattice design using data in Table 4.32.
|
Source of variation |
Degrees of freedom (df) |
Sum of squares (SS) |
Mean square
|
Computed F |
|
Replication |
1 |
981.24 |
981.24 |
2.218 |
|
Treatment (unadj.) |
24 |
9087.29 |
378.64 |
0.856 |
|
Block within replication (adj.) |
8 |
1819.56 |
227.44 |
0.514 |
|
Intra-block error |
16 |
7077.73 |
442.36 |
|
|
Total |
49 |
18965.83 |
The treatment mean square, as presented in the ANOVA table (Table 4.38), is not adjusted for block effects. As pointed out earlier, the treatment means are not free from block effects. As a result, the ANOVA does not provide a valid F-test for testing the treatment differences. Before applying F-test, the treatment means are to be adjusted for block effects and adjusted sum of squares for treatments will have to be computed. The procedure for this is given in Step 9. Though this procedure may be adopted when necessary, it is not always necessary to go through further computation unless it is indicated. For instance, in field trial with a large number of treatments, a significant difference among treatment means may generally be expected. As a less sensitive test for treatment difference, a preliminary RCBD analysis may be carried out from the results of Table 4.38.
Step 8. Preliminary RCBD analysis : The error sum of squares for RCBD analysis is obtained by first pooling the inter block error with intra-block error and completing the ANOVA table as follows.
Pooled error = Inter block error + Intra-block error (4.56)
= 1819.56 + 7077.73
= 8897.29
Table 4.39. ANOVA table for preliminary RCBD analysis.
|
Source of variation |
Degrees of freedom (df) |
Sum of squares (SS) |
Mean square
|
Computed F |
|
Replication |
1 |
981.24 |
981.24 |
|
|
Treatment |
24 |
9087.29 |
378.64 |
1.02 |
|
Pooled error |
24 |
8897.29 |
370.72 |
|
|
Total |
49 |
18965.83 |
The observed F-value of 1.02 obtained as the ratio of treatment mean square and pooled error mean square is less than the table value of F at which was 1.98 at 5% level of significance for (24, 24) degrees of freedom, and is suggestive of the fact that the treatments do not differ significantly at 5% level of significance. Since this preliminary RCBD analysis has ended up with a nonsignificant value for F, there is a need to employ a more appropriate F-test by adjusting the treatment sum of squares for block effects, since such a procedure would only increase the sensitivity by which the testing is made but not decrease it. The procedure for effecting such an adjustment to the treatment sum of squares for obtaining a more appropriate F-test for testing treatment differences is given in Step 9.
Step 9. Computation of treatment sum of squares adjusted for block effects : First, obtain the unadjusted block sum of squares within replications. Since we have already calculated the block sums B1, B2, …, B10 in Step 3, this is easily computed as follows :
Unadjusted block SS for Replication I = SSB1(unadj.)
= ![]() (4.57)
(4.57)
= ![]()
= 1219.75
Unadjusted block SS for Replication II = SSB2 (unadj.)
=![]() (4.58)
(4.58)
= ![]()
= 1850.83
Finally obtain the pooled unadjusted block sum of squares, SSB (unadj.), as
SSB (unadj.) = SSB1(unadj.) + SSB2 (unadj.) (4.59)
= 1219.75 + 1850.83 = 3070.58
Compute the following correction quantity Q to be subtracted from the unadjusted treatment sum of squares :
Q = 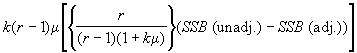 (4.60)
(4.60)
where m
= ![]() (4.61)
(4.61)
where Eb = Adjusted inter block mean square
Ee = Intra-block mean square
For our example, m
= ![]()
= - 0.189
Q = 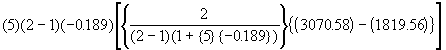
= -42989.60
Finally, subtract this quantity Q from the unadjusted treatment sum of squares to obtain the adjusted sum of squares for treatment.
SST (adj.) = SST (unadj.) - Q (4.62)
= 9087.29 - (-42989.60) = 52076.89
Set up the following ANOVA table for testing the significance of the treatment effects.
Table 4.40. ANOVA table for testing the significance of adjusted treatment means.
|
Source of variation |
Degree of freedom (df) |
Sum of squares (SS) |
Mean square
|
Computed F |
Tabular F |
|
Treatment (adj.) |
25 |
52076.89 |
2083.08 |
4.709 |
2.24 |
|
Intra-block error |
16 |
7077.73 |
442.358 |
The F-value computed for the present example has turned out to be significant at 5% level indicating that significant differences among the treatments. The sensitivity of F-test increased after eliminating the block effects. Generally, the block effect, as judged from Eb value is supposed to be greater than the intra-block error Ee. although this did not happen with the present example.
Adjustments are necessary to treatment means as well, since the ordinary treatment means are not unbiased estimates of true values of treatment means. The procedure for effecting such adjustments to eliminate block effects is as follows:
Step 10.Compute a correction term for each block by multiplying each Cb value by the quantity m ( -0.189), given by (4.61).
For Replication I, these values are :
m C1 = -8.13, m C2= -4.74, m C3 = 6.01, m C4 = -14.78, m C5 = -20.22
For Replication II, these values are :
m C6 =18.99, m C7 = 14.59, m C8 = 3.74, m C9 =1.59, m C10 = 2.95
Enter these values in the last row and last column of Table 4.34 as shown in Table 4.31. Check that the total of all m Cb values add up to zero except for rounding of error. i.e.,
m C1 + m C2 + …+ m C10= -8.13+-4.74 +…+ 2.95= 0.00
Make entries of the m Cb values for Replication I along the last column of Table 4.41 and m Cb values for Replication II along the last row of the same Table 4.41. This way of writing the correction values to be effected to the unadjusted totals for treatments will save a lot of confusion in carrying out arithmetical calculations. Each treatment total in Table 4.41 is now to be adjusted for block effects by applying the block corrections appropriate to the blocks in which that treatment appears.
Table 4. 41. Treatment totals and correction factors.
|
1 |
2 |
3 |
4 |
5 |
m C1 = |
|
260.80 |
264.60 |
266.30 |
275.90 |
284.00 |
-8.13 |
|
6 |
7 |
8 |
9 |
10 |
m C2= |
|
284.70 |
244.70 |
246.20 |
214.10 |
223.80 |
-4.74 |
|
11 |
12 |
13 |
14 |
15 |
m C3 = |
|
220.30 |
210.60 |
242.60 |
196.90 |
229.00 |
6.01 |
|
16 |
17 |
18 |
19 |
20 |
m C4= |
|
276.30 |
213.90 |
230.30 |
209.50 |
254.80 |
-14.78 |
|
21 |
22 |
23 |
24 |
25 |
m C5 = |
|
253.80 |
297.60 |
267.40 |
228.40 |
234.00 |
-20.22 |
|
m C6 =18.99 |
m C7 = 14.59 |
m C8 = 3.74 |
m C9 =1.59 |
m C10 = 2.95 |
For example, clone 1 appears in Block 1 of Replication I and Block 6 of Replication 2. Add the values m C1 and m C6 to the total for clone 1.
i.e., The adjusted treatment total for clone 1 = 260.80 -(-8.13) - 18.99 = 2.55
Since the block corrections are already entered along the row and column in Table 4.41, the adjusted treatment totals are merely obtained by the respective column and row values for m Cb in which that treatment appears. Finally, construct a table showing the treatment total adjusted for block effects. The adjusted values are shown in table 4.42 below.
Table 4.42. Adjusted treatment totals:
|
1 |
2 |
3 |
4 |
5 |
|
249.94 |
258.14 |
270.69 |
282.44 |
289.18 |
|
6 |
7 |
8 |
9 |
10 |
|
270.45 |
234.85 |
247.2 |
217.25 |
225.59 |
|
11 |
12 |
13 |
14 |
15 |
|
195.30 |
190.00 |
232.85 |
189.30 |
220.04 |
|
16 |
17 |
18 |
19 |
20 |
|
272.09 |
214.09 |
241.34 |
222.69 |
266.63 |
|
21 |
22 |
23 |
24 |
25 |
|
255.03 |
303.23 |
283.88 |
247.03 |
251.27 |
Obtain the adjusted treatment means by dividing each value by 2 since each total contains two observations from 2 replications (Table 4.43)