Abstract
Clonal selection of banana (Musa acuminata AAA, cv. 'Grande Naine') was performed under the marginal climatic conditions for banana cultivation in the Western Galilee region of Israel. The initial selection was carried out on 300 mats originating from six mother clones multiplied by meristem culture. Five clones were selected for further experiments. In the third growth cycle the total number of bunches per hectare of three out of the five selections, exceeded that of the control, while the remaining two selections performed similar or below the control. In the fourth cycle all the selections out-performed the control. Average bunch weights of two of the selected clones were significantly higher than the control. The total yield, derived from bunch weight and the number of bunches per mat of each of the selections, was higher than that of the control in both the third and the fourth cycle.
Hundred and fifty hectares of micropropagated plants derived from the selected clones were planted in the Philippines, and their performance was evaluated and compared to that of a local selection of 'Grande Naine'. The yield of the Israeli selected clones in the Philippines was significantly higher by an average value of 18 percent compared to the local clones.
Using degenerate primers, a fragment of a retro-transposable element (BR-1) that was induced by extensive in vitro culture was detected in Musa. Transcriptional activation of the banana retro-transposon was detectable by RT-PCR in the 'Long and Narrow Leaf ' mutant (LNL). The frequency of occurrence of BR-1 was analyzed in genomic DNA of the LNL mutant and in its originator non-mutant mother plant. The analysis revealed amplification of BR-1 in the mutant compared to the mother stock DNA. We postulate that retro-transposable elements are at least in part responsible for the occurrence of the LNL mutant.
1. INTRODUCTION
Cultivated bananas are natural selections originating from the center of origin of the genus Musa in the Asian Pacific region. The edible types comprise a range of natural hybrids originating from the two species Musa acuminata (A genome) and Musa balbisiana (B genome). Most cultivars are triploids (AAA, AAB or ABB genomes), parthenocarpic and sterile, though in the international trade of dessert bananas, the vast majority belongs to the AAA 'Cavendish' subgroup. Despite the intricacy of breeding infertile plants, a number of new banana cultivars have been released over the last three decades. All the new hybrids that have been accepted by growers comprise a mixture of A and B genomes. While the use of the new hybrids is rising, consumer acceptance is limited to local (not export) markets.
The main focus of banana breeding programs for the export industry is resistance to diseases and improvements of fruit quality. The extreme susceptibility of the old cultivar 'Gros Michel' to Fusarium wilt (Panama Disease) forced breeders and producers of export sectors of the industry to shift to a more resistant variety. The newly created cultivars should confer resistance or tolerance to the major diseases, in order to reduce the costs and extensive use of fungicides and nematicides. Moreover, eruption of new races of pathogens can endanger the existence of the entire banana industry, due to the narrow genetic pool of selected clones. The outbreak of viral diseases constitutes another threat to the banana industry in most production areas.
Introduction of in vitro culture allowed production of pathogen free plants and widened the genetic variability of existing cultivars, as a result of somaclonal variation [1,2,3). At the same time, due to the variation that occurs during the in vitro propagation, plants of the same cultivar selected in different locations often differ in some characteristics. Thus, the in vitro technique allows local genetic selections. An example of a banana cultivar improved by somaclonal variation was demonstrated in Taiwan by [4] who selected somaclones resistant to F. oxysporum. These clones were further improved by somaclonal variation for better field performance [5,6,7].
The high incidence of "off-types" produced by banana meristem culture is a major concern to commercial growers. At the same time, somaclonal variation is an important tool for the improvement of banana germplasm. Banana plant propagators and researchers have learned, over the years, to reduce the frequency of "off-types" to a manageable level. The majority of the banana "off-types" in the Cavendish subgroup appear as 'dwarf' or 'giant'. It has been demonstrated that both are associated with sensitivity to gibberellic acid [2]. Most types of the in-vitro generated mutations in Cavendish result from a long duration in tissue culture and from high number of cycles in the multiplication phase of the meristem culture. The specific genes responsible for these mutants have not been identified so far. Several gibberellin insensitivity mutants have been isolated and characterized in other plant species, but none of these have been associated with somaclonal variation. Unlike Cavendish types, an array of phenotypic variations was reported in plantains. Most variations in plantains were related to the morphology of the inflorescence and fruit [9]. Thus, it seems that the highly frequent types of variation are genotype specific.
The Western Galilee in northern Israel is on the border line of commercial banana production. In the summer months the solar radiation reaches very high levels, the ambient temperatures are high and rainfall is practically nil. During winter nights the temperatures occasionally drop below the freezing point [10]. Under these low temperatures banana plants are severely damaged, but nevertheless production remains adequate.
Sucker selection in Israel is practiced during the summer months and is aimed to restrict flowering to mid summer of the next year and harvest to the following winter months. Sucker selection is performed by experienced growers and is based on the number of accumulated leaves on specific dates [11,12]. The "selector" determines the number of suckers to be left for the next cycle in each mat, according to physiological conditions of the plants in the specific mat and its relations with adjoining mats. Yield is determined mainly by the number of bunches/ mat (usually 1-3 per mat) and the weight of each bunch.
In tropical regions like the Philippines, the number of bunches per mat is fixed (usually only one). Therefore clonal evaluation is influenced by bunch weight only.
Due to the relatively "harsh" climatic conditions and unique topographic location combined with strict quarantine measures, the Western Galilee is currently free of the major banana pests and diseases, including Black and Yellow Sigatoka, Banana Bunchy Top Virus, Banana Streak Virus, Panama disease and the most damaging nematode Radopholus similis [13,14]. The vectors that transmit these diseases seem unable to overcome neither the cold winter temperatures nor the low humidity in the long and dry summers.
In spite of the work reported thus far on clonal variation in banana, the genetic aspects of this phenomenon are unknown. The lack of understanding of the genetic events leading to somaclonal variation puts severe constraints on producers of tissue cultured banana plants. At the same time it opens an opportunity for selection of elite clones. In this investigation we attempted to elucidate the genetic mechanism of clonal variation that is induced by tissue culture.
2. MATERIALS AND METHODS
2.1. Initial evaluation
The initial selection was performed on 300 mats of 'Grande Naine' produced from six mother clones, propagated by meristem culture. The in vitro cultures were initiated from apical meristem explants and were propagated and rooted according [15]. Hardening, potting and growth prior to transfer to the field were carried out as practiced commercially by Rahan Meristem. After three months of ex vitro hardening, the plants were planted in the Western Galilee Banana Experimental Station.
Mats were arranged in wide "tram-line" paired rows [16]. The distance between each pair was 6.0 m and between rows within the pair 3.0 m. The planting distances within the row were 2.7 m. Thus, the total density was 832 mats/ha. Drip irrigation, fertilization and all other agrotechnical procedures were according to standard methods practiced in the Western Galilee.
The evaluation was based on data collected from each individual mat over a six year period. The criterion for selection was yield per mat, calculated by adding the weight of the bunches produced in the mat. Four clones, '5-1', '6-6', '37-5' and '42-5', which performed exceptionally well, were selected for further evaluation. A fifth clone '17-1' was selected separately in response to cold resistance characteristics (data not shown).
2.2. Clonal evaluation
Based on the results of the above experiment, the best clones were multiplied by meristem culture as mentioned above.
Seventy plants of each of the selected clones and of a control representing the average value for all 300 original mats were used for the experiment.
The planting material consisted of hardened micropropagated plants, three months after hardening. Planting distances were as mentioned above (823 mats /hectare). The plants were arranged in randomized blocks of five replicates. The numbers of bunches/ mat and bunch weights were recorded for 10 mats in each replicate (excluding the mats at the end of the rows). Altogether 20-30 bunches per replicate and over 120 bunches per clone were evaluated each year.
In the winter following planting temperatures were unusually low, hindering plant growth for two consecutive cycles. The yield in the first two years was unusually low and as a consequence the results of these cycles were ignored. The data presented in this report include the results of the third and the fourth years after planting.
Approximately 30 hectares of micropropagated plants of the selected clones were planted in a commercial banana plantation in the Western Galilee region. These 30 hectares served as a source of explants for the evaluation experiments in the Philippines. The parameters for choosing the explants within the plant population were flowering time and yield.
In the Philippines the performance of the Israeli selected clones was compared to the leading 'Grande Naine' selection, which was introduced to the Philippines from Ecuador (Ballesteros, personal communication). The local plants were also propagated by meristem culture and potted as mentioned above.
Planting scheme in the Philippines was in "hedgerows" 4.5m apart [16]. The plants were arranged within the row in a triangular configuration at distances of 1.25m. Total density was 1973 plants per hectare.
The plants were drip irrigated and fertilized according to standard methods practiced in the Philippines. The experiment was conducted in 3 separate farms, each containing both Israeli and Ecuadorian selections at a ratio of 1:1, in 50 hectares plots. The values presented are the calculated means of all three plots.
2.3. Characterization of 'off-types'
2.3.1 Induction of variation by extensive duration of meristems in in-vitro conditions
Meristem culture was performed on a single 'Grande Naine' clone employing a standard protocol used at Rahan Meristem. The meristems were sub-cultured extensively (23 cycles) to induce somaclonal variations. The in vitro plantlets were transferred to a stage III medium containing 10 ppm of GA3. Analysis of GA sensitivity was examined by measuring elongation and the distance between internodes of the in vitro plants following a four weeks culture period on the GA enriched medium [2].
Further analysis of somaclonal variation was carried out in ex-vitro conditions. Following acclimatization the plants were planted in 5 L pots and placed in a poly-ethylene covered greenhouse for approximately 16 weeks prior to selection of 'off-types'. Candidate plants showing distinct somaclonal variations were selected and used for further analysis. Stability of the different mutations was monitored following the growth of the selected clones in a commercial plantation in Rosh Hanikra, Israel.
2.3.2. DNA isolation and Southern blot analysis
Samples (2.5 g) of fully expanded leaf blade tissue were harvested and ground by mortar and pestle under liquid nitrogen. The samples were homogenized in 25 mL extraction buffer containing 4% (W/V) CTAB, 10 mM Tris-HCl pH 8, 1.4 M NaCl and 20 mM EDTA. The extracts were placed at 650C for 30 min. After cooling to room temperature an equal volume of chloroform:isoamyl alcohol (20:1) was added and after 15 min. of incubation at room temperature the mixture was centrifuged at 5000 rpm for 5 min. The supernatant was filtered through 5 layers of cheesecloth and an equal volume of ice cold iso-propanol was added to the filtrate. Following addition of NaCl to a final concentration of 0.1 M the samples were kept at -200C for one hr. and subsequently centrifuged for 15 min. at 11,000 rpm at 40C. The resulting pellet was resuspended in 3 mL of 70% ethyl alcohol, the mixture was centrifuged as above, and the resulting pellet was resuspended in 0.5 mL distilled water. Aliquots of ten µg DNA were digested with EcoRI and separated on a 1.2% agarose gel, stained with ethidium bromide and blotted onto a Nytran membrane. Transfer of DNA and hybridization was according to [17]. The membranes were probed with a 344 bp PCR product previously isolated from banana genomic DNA using Tos-1 primers [18].
2.3.3. RNA isolation and RT PCR
Five grams of young leaf tissue were ground in liquid nitrogen and homogenized with extraction buffer containing 0.2 M Tris-HCl pH 8.5, 0.33 M LiCl2, 10 mM EDTA and 1% PVPP. The homogenate was centrifuged for 20 min. at 7000 rpm. Following centrifugation the supernatant was filtered through five layers of cheesecloth and subsequently cold ethanol to a volume of 10% (V/V) and 3% (V/V) 3.3 M NaAc was added. The mixture was centrifuged at 10000 rpm for 10 min. and the supernatant collected and extracted twice with an equal volume of phenol: chloroform and once in chloroform. The aqueous phase was precipitated over night at -200C in an equal volume of isopropanol and 1/10 volumes of 3.3 M NaAc (pH 6.1). Following centrifugation at 7000 rpm at 40C for 20 min. the pellet was resuspended in cold 80% ethyl alcohol, centrifuged as above and finally rinsed in absolute alcohol. The pellet was resuspended in DEPC treated distilled water to a concentration of 2 µg per µL.
The RT reaction mixture (total volume 25 µL) contained 0.5 µg RNA, 1 mM (each) of a nucleotide mixture, 40 pmoles of oligo T18 primer, 50 units of RNA's inhibitor, 20 units of RT enzyme (AMV - Boehringer Mannheim) and 1 µL of enzyme reaction buffer. The reaction mixture was incubated for 10 min. at 250C, 60 min. at 370C and 5 min. at 950C.
PCR was performed in a 50 µL reaction mixture containing: a 1 µL aliquot from the RT reaction, 0.4 mM (each) nucleotide mixture, Tos- 1 upper and lower primers (50 pmoles each) and 2.5 units of Taq-polymerase (KlenTaq). The reaction was performed at 950C dissociation for 2 min. followed by 25 cycles of 30 sec. at 950C, 60 sec of annealing at 600C, 1 min. of primer extension at 720C and finally the mixture was kept at 720C for an additional 7 min. Following the PCR the samples were separated on a 1.2% agarose gel, stained with ethidium bromide and visualized with a UV lamp.
3. RESULTS
3.1. Initial selection
Out of the 300 mats that were planted in 1984, five clones performed substantially better than the control (combined value of all the clones used for the selection). The yield of each of the selected clones exceeded the control (Table 1).
Table 1 Average yield of 5 selected clones in the 6 years of the experiment, as compared to control. Yield calculated according to a density of 832 mats/ha
|
Parameter |
Clone |
|||||
|
6 - 6 |
5 -1 |
17- 1 |
42- 5 |
37- 5 |
Control |
|
|
Bunch wt. (kg) |
37.47 |
37.82 |
33.44 |
37.44 |
35.68 |
30.47 |
|
Bunches/ mat/ year |
2.83 |
2.83 |
2.33 |
2.83 |
3.00 |
1.74 |
|
Calculated yield (t/ha/yr) |
87.38 |
88.19 |
64.22 |
87.31 |
88.10 |
43.59 |
Five selections '5-1', '6-6', '37-5', '42-5' and '17-1' generated 35-75 % more bunches per mat compared to the average (control). The average bunch weight of all five selections exceeded the control by 2.9-7.3 kg (9.5-24%). As a consequence, the yield of the four selections was approximately twice as high as the control, while the yield of selection '17-1' was only 50% higher (Table 1).
3.2. Evaluation of selected clones under Israeli conditions
The total number of bunches/ha (Figure 1a) of clone '17-1', '42-5', and '37-5' in the third year of production exceeded the control ('42-5' not statistical significant), while clones '6-6' and '5-1' were below or similar to the control. In the fourth cycle (Figure 1b), all the selections performed better than the control, though only in clone '17-1' the difference in the number of bunches was statistically significant (P=0.05). Clone '6-6' generated the lowest number of bunches in the third cycle (1742 compared to 1753 in the control), but at the fourth cycle it out-performed the control by approximately 10%. Selection '37-5' generated the highest number of bunches per hectare in both growth cycles (more than 11% higher than the control).
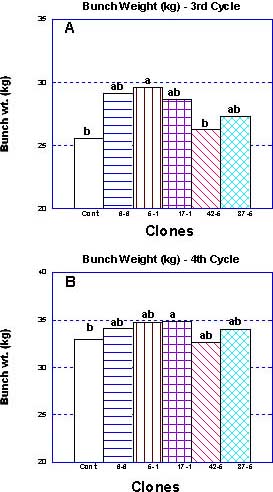
Average bunch weights of selections '5-1' and '17-1' were significantly (P= 0.05) above the control value in both the third and fourth cycles (Figure 2 a,b). On the other hand, clone '42-5' generated the least weight per bunch in both cycles.
Figure 1 Number of bunches per hectare for the selected clones and for the control population during the third (A) and fourth (B) production cycles in the Western Galilee, Israel. Measurements are means of 50 mats (>120 bunches). Mean separation by multiple range test P=0.05
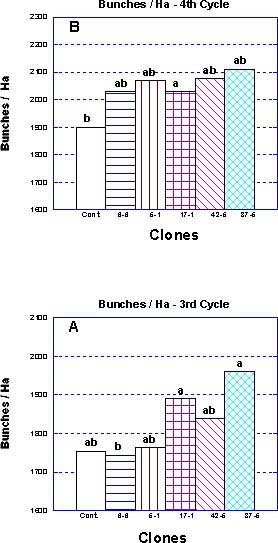
Figure 2 Bunch weight (Kg) of fruits harvested from the selected clones and from the control population during the third (A) and fourth (B) production cycles in the Western Galilee, Israel. Measurements are means of 50 mats (>120 bunches). Mean separation by multiple range test P= 0.05
The total calculated yield, derived from the number of bunches and bunch weight, was higher in each of the clones compared to the control. The ratio between the selections followed a similar pattern (Figure 3).
3.3. Comparison of selected clones with a local selection in the Philippines
Although the values for bunch weight and box/stem ratio (Figure 4,5) for Israeli selections were consistently higher throughout the 20 months of the experimental period, all selections followed a similar pattern of temporal changes in fruit production.
Figure 3 Yield in tons/ha of the selected clones and of the control population during the third (A) and fourth (B) production cycles in the Western Galilee, Israel. Measurements are means of 50 mats (>120 bunches). Mean separation by multiple range test P=0.05
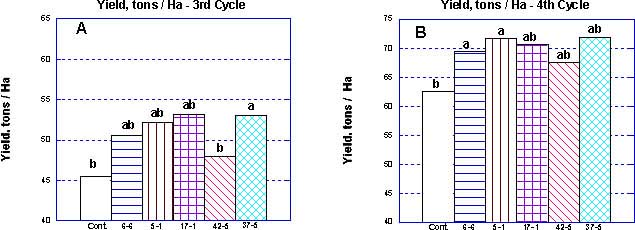
Figure 4 Bunch weight (kg.) of "R" selection (a mixture of 5 Israeli selected banana clones of cv. 'Grande Naine') compared to local clones ("L" selection) over a 20 months period, starting at the second year of production in the Philippines. Each point represents the means (±SD) of three independent replications of about 50 hectares each
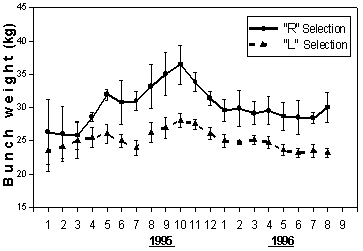
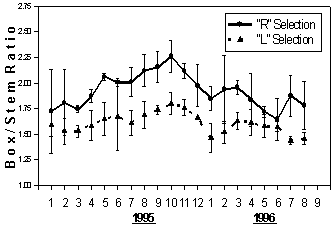
Figure 5 Box / stem ratio of "R" selection (a mixture of 5 Israeli selected banana clones of cv. 'Grande Naine') compared to local clones ("L" selection) over a 20 months period, starting at the second year of production in the Philippines. Each point represents the means (±SD) of three independent replications of about 50 hectares each
Bunch weight of both selections gradually rose from January 1995 and reached maxima in October (Figure 4). The weight fell during 1996 in both selections. A wide and significant difference in favor of the Israeli selection appeared from April 1995 and on.
The difference reached a peak of over 30% (8.5 kg/bunch) and moderately declined in the subsequent months to approximately 5 kg/bunch (20%).
Box to stem ratio, which reflects both quality and yield, was higher for the Israeli selection compared to the local one (Fig. 5). The difference gradually increased from January to October 1995 and slowly declined until very small differences were noted in the middle of the second year of measurements. The highest value of 2.26 and 1.8 boxes/ stem respectively for the Israeli and local selection was reached in October 1995.
3.4. Characterization of common somaclonal variants in mass propagated banana plants
Somaclonal variants with both high and low sensitivity to GA were generated as a consequence of an extensive duration in tissue culture. Both 'off-types' were detectable by a relatively simple bioassay developed in our laboratory (Table 2). GA sensitivity differed significantly between the different plants. The dwarf genotype had approximately 40% shorter internodes in comparison to the normal phenotype, but showed sensitivity to GA. On the other hand the 'Extra Dwarf' phenotype was insensitive to the presence of GA in the medium.
Table 2 Influence of GA3 on "internode" length (cm) of normal in-vitro and mutant banana Grande Naine plantlets. Plantlets were grown for 5 weeks on GA-containing or hormone-free media
|
Clone / Treatment |
Normal |
Dwarf |
Extra dwarf mutant |
|
Control (hormone free) |
*) |
10.8 c |
9.6 d |
|
GA3 (10 mg/L) |
21.7 a |
14.9 b |
9.6 d |
* Values indicated by different letters represent statistical significance (P=0.05) using a multiple range variant analysis.
Figure 6 Somaclonal variation. Leaf blades of two 'Cavendish' clones produced from the same mother explant. The upper leaf (LNL) represents the Long and Narrow Leaf mutant, the lower leaf (WT) is Wild Type

Another common variation is the phenotype of long and narrow leaf (Figure 6). This phenotype is also known as 'Giant' by growers.
3.5. Detection of retro-transposons in banana
We have attempted to detect expression of retro-transposable elements in leaves from Dwarf, Extra Dwarf and LNL plants that were induced to transcribe retro-elements by extensive duration of tissue culture. After twelve subcultures leaf samples were analyzed for the presence of mRNA encoding retro-elements. Using a specific set of primers we were able to detect a 300 bp retro-element in the LNL mutant (Figure 7).
Figure 7 PCR (panel A) and RT-PCR (panel B) products using TOS 17 primers to amplify Ban-Retro 1. Lanes 1-5 in panel A represent PCR products from DNA samples of plantlets at different duration periods in tissue culture. Lanes 7-12 in panel B represent different RNA samples used for RT-PCR with (+) or without (-) RT enzyme (control). Lane 6 - molecular marker DNA (Hind III cut Phi-X)
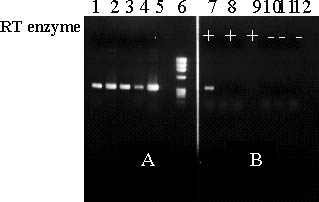
The retro-element was only detected in the presence of the RTase in the synthesis of the cDNA and was not observed in any of the six wild type plants.
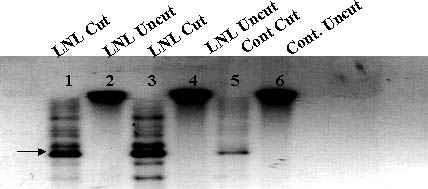
DNA gel blot analysis was used to estimate the complexity of retro-transposon enrichment of banana genomes following activation by tissue culture. Additional bands appeared in the lane loaded with the retro-activated mutant (LNL) DNA compared to that of the wild type (Figure 8). Furthermore, at least two bands that were present in the control show a significant higher label intensity as compared to the wild type.
Figure 8 Southern hybridization of control (lanes 5,6) and LNL mutants (lanes 1-4) genomic DNA with 32P labeled Ban-Retro 1 probe. Ten µg DNA were either uncut with EcoR1(lanes 2,4,6) or cut (lanes 1,3,5)
4. DISCUSSION
Components of yield in bananas include number of bunches per mat, weight of each bunch and number of mats per hectare, which together determine the potential production in tons per hectare. In tropical areas, where plants grow continuously, the length of each growth cycle is an additional important component of yield, while in the Western Galilee, Israel, growers can only expect one growth cycle per year. In order to achieve high yield and better protection from adverse climatic conditions, producers tend to increase plant density by increasing the number of bunch bearing stems in each mat [11,19]. Another important factor worth considering (mainly for exporting producers) is fruit quality, with emphasis on finger size and appearance of the fruit. Since yield has a positive correlation with all the parameters used in our initial selection, we predicted that selection for total yield in Israel would be useful in the tropics as well.
Although a narrow genetic base was sampled in our initial selection program, the quantitative and qualitative parameters were manifested in the subsequent field trials. The five clones selected performed above the mean values measured for the total population in all the relevant parameters: number of bunches per mat, average bunch weight and total yields per hectare (Table 1).
The second step of clonal evaluation was performed on more than 120 bunch bearing plants per year, of each of the selected clones. The achievements of these clones in total yield, though differing from each other, were consistently above the control population (Figure 1a,b Figure 2a,b).
The difference in performance between the third and fourth cycle can be attributed to the difference in climatic conditions. During the winter season that preceeded the third cycle, the temperatures measured in the Western Galilee were unusually low. The residual effects of the harsh climate resulted in lower yields than expected for all clones including the control (Figures 2a,b). However, even under the sub-optimal conditions of the third cycle, clones '17-'1 and '37-'5 produced approximately 17 percent more fruit per hectare compared to the control population (Figure 3a). The traits that mainly contribute to yield are bunch weight and the number of bunches per hectare. Our data reveal a significant difference between the clones, in the context of these two parameters. While bunches of clones '6-6' and '5-1' weighed more than '42-5', the number of bunches per hectare for '42-5' out-performed the others in both the third and fourth cycle of growth (Figure 1,3). It is likely that a mixed population of several selected clones will be advantageous in a different ecological environment. Furthermore, to avoid deleterious effects due to a mono-culture population, we combined all five clones for evaluation in the Philippines. Propagation by meristem culture allowed an even mix of the clonal selections.
A crucial parameter for the banana export industry is fruit quality. Fruit loss due to inferior quality can reach 40 percent in some cases. To include this important characteristic in our evaluation, the yield for the Philippines was expressed as "box/stem ratio" in addition to "bunch weight" (rather than fruit weight per hectare). This ratio was significantly higher for the plants selected in Israel compared to locally selected clones (Figure 5). The difference reached an average value of 18 percent for the entire evaluation period and peaked during the most productive period around October 1995. Similar results were achieved with bunch weight (Figure 4).
The genetic clonal divergence supports the presumption of instability of the banana genome [3]. Given that both clones (the one selected in the Philippines and that selected in Israel) clearly differed in various components of yield, shows that selection is influenced by environmental conditions. The geographic location of the Western Galilee allows selection unmasked by the effects of diseases and pests. Our data indicate that selection in the ecological conditions of the Western Galilee provides better results in ecosystems that involve biotic pressures. However, after several cycles of propagation, the Israeli clones seem to lose their advantage (unpublished data), probably due to a genetic drift caused by biotic pressure.
The limited gene pool of cultivated bananas for the export industry has led researchers to use alternative methods of germplasm improvement such as mutation breeding [20] and recombinant DNA technology [21,22]. However, since both of these techniques rely on small genetic changes, normally single genes, it is crucial to start with a good genetic baseline and with plants free of viruses. In this regard, our data suggest that pre-selected clones in marginal climates for banana production are advantageous.
Interestingly, the occurrence of "off-types" following extensive tissue culture is higher than expected from random cell mutation. However, most detectable 'off-types' exhibit either higher or lower sensitivity to GA (Table 2). The results obtained by the GA assay on in vitro plantlets provide evidence to the hypothesis that 'Dwarf' and 'Giant' phenotypes are related to GA sensitivity. [2] have shown that in addition to the difference in sensitivity, partitioning between the different GA metabolites differed between 'Dwarf', 'Giant' and the normal phenotypes.
3.7. Involvement of retro-transposons in somaclonal variation
The high frequency of deviation from the original clone, occurring during the in vitro culture, suggests that the tissue culture process promotes genomic changes. However, the mechanism of somaclonal mutations in bananas is unknown. Recent studies [18] provided clear evidence that extensive duration of rice cell culture activated retro-transposing elements. Furthermore, the tobacco retro-element Tto 1 undergoes transcriptional activation by wounding as well as by methyl jasmonate [23]. The structural features as well as transcriptional activation of retro-elements resemble retroviruses. Under normal conditions they remain dormant and upon activation they are transcribed, reverse transcribed to cDNA molecules and reintegrated in new loci in the genome.
The DNA of the pre-cultured meristem and the mutants was analyzed by Southern hybridization using BR-1 as a probe. The probe hybridized to two fragments in the control (Fig. 7), while the LNL mutant hybridized to at least four additional fragments. The addition of bands on the Southern blot indicates a propagation of the retro-element in the LNL mutants. At this point, the number of integrated copies represented per band is unclear. However, in the LNL the hybridized signal was intensified. This may indicate multiplication of retro-elements in a close proximity to the original retro-sequence. A cascade form of retro-elements has been reported in other species [24]. This phenomenon may explain the high frequency of a single phenotypic variation. If a gene associated with GA sensitivity resides in close proximity to the original retro-sequence and the distribution of insertions is biased to short distances, we expect a high rate of mutants involving GA sensitivity. We are currently investigating this hypothesis.
5. REFERENCES
[1] DANIELLS, J.W. and Smith, M.K. 1992. Somatic mutations of bananas, their stability and potential. p. 162-171. In: Recent developments in banana cultivation technology. Proc. International Symp. Chiuju, Pintung, Taiwan 14-18 Dec. 1992. TBRI/ INIBAP/ ASPNET.
[2] REUVENI, O., Israeli, Y. and Lahav, E. 1996. Somaclonal variation in bananas. pp. 174-196. In: Bajaj, Y.P.S. (ed). Biotechnology in agriculture and forestry, somaclonal variation in crop improvement II.
[3] WALTHER, R., Ilan, A., Lerer, A., Duvdevani, A. and Khayat, E. 1997. Analysis of somaclonal variation in tissue cultured banana plants (Musa AAA cv. 'Grande Naine'). Acta Horticulturae 447.
[4] HWANG, S.C. and Ko, W.H. 1988. Mutants of Cavendish banana resistant to race-4 of Fusarium oxysporumf.sp. cubense. Plant Protection Bulletin 30: 386-392
[5] HWANG, S.C. and Ko, W.H. 1990. Selection of improved Cavendish banana mutants resistant to race-4 of Fusarium oxysporumf.sp. cubense. Acta Horticulturae 275:417-423.
[6] HWANG, S.C. and Tang, C.Y. 1995. Somaclonal variation and its use in Taiwan. (Paper resented to MARDI, International banana workshop: New frontiers in resistance breeding for nematodes, fusarium and sigatoka. Serdang, Malaysia 2-5 Oct. 1995)
[7] SMITH, M.K., Hamill, S.D., Langdon, P.W. and Pegg, K.G. 1993. In vitro mutation breeding for the development of bananas with resistance to race-4 fusarium wilt (Fusarium oxysporumf.sp. cubense). FAO/IAEA. Proc. 1st research co-ordination meeting. In vitro mutation breeding of bananas and plantains. Vienna, Austria 29 May-2 June 1989. p. 66-78.
[8] COTE, F., Sandoval, J., Marie, Ph. and Auboiron, E. 1993. Variations in micropropagated ananas and plantains. Literature Survey, Fruits 48: 11-20.
[9] VUYLSTEKE, D., Swennen, R. and De Langhe, E. 1991. Somaclonal variations in plantains (Musa spp., AAB group) derived from shoot-tip culture. Fruits: 46 (4): 429-439.
[10] KATZENELSON, Y. 1985. Climatic conditions in Israeli Regions. p.189-235. in Arnon, I. (ed) Encyclopedia of Agriculture. Volume 1. (in Hebrew). Agr. Enc. Publ.
[11] ZIV, D. 1962. Researches on the banana and irrigated crop cycle. (in Hebrew) "Hassadeh" publication.
[12] STOLLER, S., Ziv, D., Smueli, E., Zuta, Z. and Schneidmesser, B. 1952. Banana researches. (in Hebrew). "Hassadeh" publication.
[13] PELED, A. 1976. The banana. p.225-230. in Halperin, H. (ed) Encyclopedia of Agriculture. Volume 3. (in Hebrew). Agr. Enc. Publ.
[14] COHN, R. 1987. Catalog of plant diseases in Israel. Publication of the Volcani Center, Agricultural Research Organization.
[15] CRONAUER, S.S. and Krikorian, A.D. 1984. Multiplication of Musa from excised stem apices. Annals of Botany 53: 321-328.
[16] ROBINSON, J.C. Systems of cultivation and management. 1995. pp. 35-36. In: Gowens (ed.) Bananas and plantains. Chapman & Hall.
[17] SAMBROOK, J., Fritsch, E. F., and Maniatis, T. olecular Cloning, A laboratory Manual. Cold Spring Harbor NY, Coldspring Harbor Laboratory Press.
[18] HIROCHIKA, H., Sugimoto, K., Otsuki, Y., Tsugawa, H. and Kanda, M. 1996. Retrotransposons of rice involved in mutations induced by tissue culture. Proc. Natl. Acad. Sci. USA 93, pp. 7783-7788.
[19] OPPENHEIMER, H. and Gottreich, M. 1959. Researches on development of the banana in the coastal region. Israeli Ministry of Agriculture. Beit-Dagan Agr. Res. St. Report 9(3-4): 229-245. (in Hebrew).
[20] NOVAK, F.J., Afza, R., van Duren, M. and Omar, M.S. 1990. Mutation induction by gamma irradiation of in vitro cultured shoot tips of banana and plantain (Musa cvs.). Tropical Agriculture (Trinidad) 67(1): 21-28.
[21] MAY, G.D., Rownak, A., Mason, H., Wiecke, A., Novak, F.J. and Arntzen C.J. 1995. Generation of transgenic banana (Musa acuminata) plants via Agrobacterium mediated transformation. Biotechnology 13: 486-492.
[22] SAGI, L., Panis, B., Remy, S., Schoofs, H., deSmet, K., Swennen, R. and Cammue P.A. 1995. Genetic transformation of banana and plantain (Musa spp.) via particle bombardment. Biotechnology 13: 481-485.
[23] TAKEDA, S., Sugimoto, K., Otsuki, H. and Hirochika H. 1998. Transcriptional activation of the tobacco retrotransposon Tto 1 by wounding and metyl jasmonate. Plant Molecular Biology 36: 365-376.
[24] SANMIGUEL, P. et al. 1996. Nested retrotransposons in the intergenic regions of the maize genome. cience 274: 765-768.
| [9] Rahan Meristem Dept. of
Research and Development Kibbutz Rosh Hanikra, 22825 Western Galilee, Israel. [10] Rahan Meristem Dept. of Research and Development Kibbutz Rosh Hanikra, 22825 Western Galilee, Israel. [11] Department of Fruit Trees, Agricultural Research Organization The Volcani Center Beit-Dagan 50250, Israel. [12] Marsman - Drysdale Group, Santos, National Highway Polomolok South Catabato The Philippines. |