Rahan Meristem Ltd
Dept of R&D
Kibbutz Rosh Hanikra
22825
Israel
Abstract
The 3' end of expressed sequence tags (EST's) cDNA from Musa acuminata are deposited in two databases (db). One db consists of transcripts expressed during fruit maturation and the second is a collection of cDNA's from leaves of the diploid variety Calcutta 4 that show differential expression in comparison to the Cavendish idio-type. The cDNA's deposited in the databases were clustered into nonredundant groups and annotated according to homology prediction using BLAST algorithms. The entire databases are periodically compared by BLAST to the NCBI public protein databases as well as to a private Arabidopsis transcriptom database. Candidate genes, postulated to be involved in fruit ripening/senescence or to be related to disease resistance signaling or execution of the defense response, were analyzed for differential expression by Northern hybridization.
1. INTRODUCTION
Development of stable and reproducible transformation and regeneration technologies opened new horizons in banana and plantain breeding. Several transformation strategies have been published in the last eight years by different banana biotechnologists [1-5]. Despite technical difficulties of transforming a monocot species, transformation protocols are readily available for most Musa cultivars.
After completion of the Arabidopsis and rice genome draft sequence, high expectations were created for functional annotation of genes and their products of the major crop plants, including Musa. To live up to such expectations will be a tall order, because considerable obstacles remain to be surmounted, but this is a worthwhile challenge that ought to be taken on. The main obstacle is the assignment of function to genes, particularly the effect of specific genes on disease resistance in a resistant background or involvement of genes in ripening and senescing processes. Fortunately, this issue is being ameliorated using enhanced understanding of the evolutionary history of genes, as inferred from detailed sequence comparisons. Homology, the evolutionary descent of genes from a common ancestor provides vital evidence in the prediction of molecular function. However, in order to use bioinformatic data for gene annotation, a considerable volume of raw information regarding sequences and expression characteristics must be deposited in an organized manner. Once at hand, homology relation to annotated entries in public databases may be utilized for prediction of gene function.
Despite growing interest in banana biotechnology, the pool of Musa genes in public databases is relatively small (of the approximately 300 accessions placed in the NCBI database, less than 25% are annotated cDNA's). This low number of annotated cDNA clones from Musa hampers the prospect of discovering genes via a homology search analysis of expressed transcripts of partial known sequences (ESTs). An extensive enrichment of the EST data is indispensable in order to predict functional genes by BLAST against the Musa genome. Given that a Musa genome initiative was recently instigated, the need for a large cDNA collection becomes essential. To this end Rahan Meristem has launched a limited functional genomic project of cDNA clones isolated from Musa acuminata fruit at different stages of development as well as clones that represent genes involved in defense mechanisms against invading pathogens. In the present report we reveal the methodology used and preliminary data.
Fortunately, the two commonly cultivated Musa species have relatively small genomes. The genome sizes are estimated to be 537 Mbp and 613 Mbp respectively for M. balbisiana and M. acuminata [6]. The size is somewhat larger than rice (430 Mbp) and Arabidopsis (125 Mbp) [7], but still smaller than most known higher plant species.
2. INSERTIONAL MUTAGENESIS FOR GENE TAGGING AND PROMOTER TRAPPING
Insertional mutagenesis using T-DNA or transposable elements has provided many useful mutations in crop plants [8-10]. Libraries of mutants are generated and screened for specific or visual phenotypes. In most genomic projects where transposon gene tagging is employed an Ac/Ds combination is inserted in transgenic lines. The Ac is an autonomous element containing a functional transposase enzyme whereas the Ds element is aberrant. The maize Ac and Ds elements are both active in heterologous systems. After several cycles of transposition plants are cross-hybridised with Wild Type (WT) plants and progenies are subsequently segregated. Mutants containing only the Ds insert are later screened for expression of detectable phenotypes. Ds mutants devoid of the Ac insert are stable while the Ac containing segregants continue to transpose and hence are unstable mutants.
Given the lack of ability of triploid Musa cultivars to cross hybridize, there is no possibility to partition the Ac element from the Ds in a co-transformed plant. On the other hand, the transposition of Ac may be indeterminate as long as the transposase enzyme remains active. As a consequence transgenic lines containing Ac will give rise to chimeric plants with respect to the locus of insertion.
We have introduced the maize Ac transposable element into the Musa genome and followed excision and insertion of the element in numerous transgenic lines. The main purpose was to determine the behavior of the maize Ac in Musa with respect to frequencies of transposition and distribution of insertions along chromosomes. The constructs we have used include an Ac element fused to GUS reporter under the 35S promoter (Figure 1), kindly provided by Professor A. Levi (Weizmann Institute Israel). The standard transformation protocol of the scalp system was applied to introduce the Ac element [2]. PCR analysis of a variety of mutants revealed that most carried a chimeric pattern with regard to expression of the foreign genes. Accordingly only a few transgenic lines (tissue cultured siblings) showed clear differences in the banding pattern on Southern hybridization blots (data not shown). Nevertheless, the majority of regenerated plants (approximately 90% of the transgenic lines) showed excision of the transposable element. Attempts were made to stabilize the Ac element following a limited number of transpositions, by silencing the gene encoding the transposase enzyme after excision (Fig. 1). In most plants expressing the antisense construct the Ac element was excised at least once. This approach appeared adequate for transposon mutagenesis in non-fertile genotypes. Other options of post transcriptional gene silencing (PTGS) are now available and would be expected to be more effective for stopping transpositions soon after the transformation event. Double stranded RNA constructs in which the sense and antisense of the same sequence are cloned consecutively but separated by an intron (RNAi), appears to be a powerful silencing tool in plant cells [11]. The RNAi technology should be further evaluated in a one-element transposon mutagenesis. A bank of mutated banana clones, preferably of a diploid genotype, would be an invaluable asset to breeders and biotechnologists. Insertion mutagenesis (both by T-DNA or transposons) could be kept for a long duration in tissue culture in minimal conditions or cryo-preserved.
3. MUSA EST GENEBANKS AND BIOINFORMATIC ANALYSIS
cDNA libraries were constructed at Rahan Meristem from fruit in different stages of ripening and senescence, leaves (young and old) from two genotypes of Musa acuminata (Calcutta 4 and Cavendish) and from leaves infected with the fungal pathogen Mycosphaerella fijiensis. EST's and full-length cDNA clones are currently being characterized and deposited in a Musa database, using the MySQL platform. A subset of clustered EST's and full-length cDNA's of interest were analyzed for differential expression by RNA blot hybridizations. In order to facilitate in-depth analysis of the collected sequences a private database is a vital tool. Database (db) design and normalization for the development of specific prediction analysis are invaluable. In this context, a desktop based database software was generated. All entries in the db are analyzed on a monthly basis by BLASTX against public databases. Both MySQL and MS Access are easy to manage and may be used with MS Windows interface. Amino acid similarities detected in a sequence are compared to the NCBI protein db and to SWISSPROT, then quantified in an alignment score S using a local alignment algorithm, gap penalties and a substitution matrix [12]. From the above score an E value is calculated. The assigned value integrates an alignment of a number of hits that scored equivalent to, or better than the S value that are expected in a random sequence from the database. Thus, an alignment with an E value of 1 is considered not to be significant, 0.1 value is somewhat significant and 0.01 is highly significant. All annotations in the dbs are based on a score of E=0.1 or better.
Figure 1 Constructs used for transposon mutagenesis.
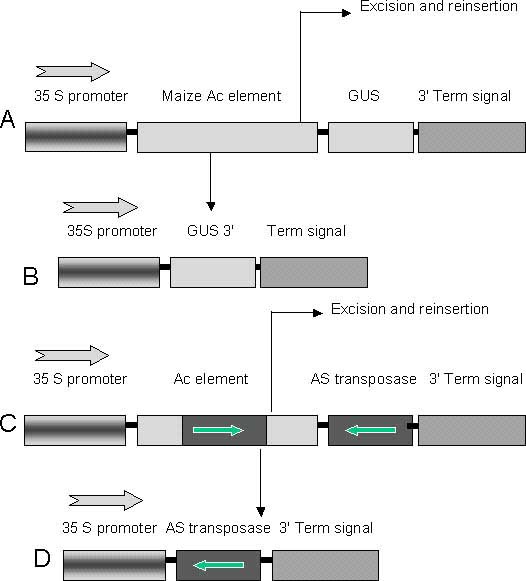
(A): Construct containing the Cauliflower 35 S promoter and the maize Ac element interrupting GUS reporter sequence. (B): Upon excision of the Ac element in the transgenic plants the GUS gene is expressed. (C): Construct containing the Cauliflower 35 S promoter and the maize Ac element and interrupting antisense expression of the transposase transcript. (D): Upon excision of the Ac element in the transgenic plants the antisense transcript is expressed. Arrows above the promoter indicate direction of transcription, smaller arrows in the boxes represent orientation of the transcript and the arrows indicated by fine lines represent excision of DNA
4. GENES INVOLVED IN FRUIT RIPENING AND SENESCENCE
The process of fruit ripening entails activation of regulatory genes that precedes the execution of the climacteric phase. The transition from pre-climacteric to the climacteric stage is triggered by a sharp rise of ethylene synthesis and is followed by an elevated rate of respiration [13]. Despite the dramatic metabolic changes occurring after the onset of the climacteric stage, the process is highly regulated rather than being chaotic [14]. The sudden metabolic transition is dictated by activation of a battery of regulatory and executing genes. Changes in the profile of mRNA and protein are the first signs of the phase transition [15]. Several fruit specific genes are up-regulated following a burst of ethylene during the process of fruit ripening. An inventory of cDNA clones isolated from fruit tissue libraries was determined in several species including tomatoes [16], strawberry [17] and cucumber [18] to name a few. The clones were categorized according to temporal parameters; response to ethylene and genotype specificity.
Differentially expressed genes, which are activated in the post climacteric phase of fruit development, were analyzed in the peel and pulp of banana. Using suppression subtractive hybridization (SSH) we have isolated over 300 partial cDNA's encoding genes, which are expressed during the final stages of fruit development (senescence). High throughput screening by membrane hybridization was employed for preliminary selection of candidate genes involved in regulation of the onset of senescence (shown in Figure 2, Table 1).
Sequence analysis and blasts against GeneBank databases revealed approximately eighty non-redundant clones, which were upregulated in the post-climacteric phase. Most, but not all of these genes were upregulated in the green fruit after a 24 hr exposure to 1,000 ppm of ethylene. The pool of sequenced upregulated cDNA's fall into one of three major functional categories:
Genes involved in metabolic processes, mainly carbohydrates and lipid components
Genes involved in cellular regulation (protein kinases, transcription factors etc.)
Genes involved in protection from pathogens and environmental stress conditions - metallothionein like protein, thaumatin like protein, super oxide dismutase, osmotin-like protein, pathogen related (PR) proteins, etc.
A significant number of sequences showed no substantial homology to functional genes in the GeneBank. Part of the unidentified sequences have distinct DNA binding domains suggesting that these genes are involved in regulation of gene expression. Several hits of Myb and homeotic box domains (mainly carpel related transcription factors) were identified in the pool of fruit cDNA.
Interestingly, the expression of various genes, particularly those associated with senescence, showed a different expression pattern in fruit of different genotypes. A distinction can be made in this context between the contribution of the A and B genomes.
Figure 2 Northern blot analysis of banana (Musa acuminata cv. Cavendish) fruit transcripts probed with cDNA from post-climacteric fruit subtracted from pre-climacteric fruit (as detailed in Table 1). Total RNA (15 µg per lane) were loaded in each lane of a 1% agarose gel and blotted onto a nylon membrane. Lane 1: preclimacteric stage (day 0 after harvest-green fruit), lane 2: climacteric stage (day 2-3 after harvest -color break stage), lane 3: post climacteric stage (days 6-7-ripe yellow fruit), lane 4: senescing fruit (days 10-12 after harvest -fruit showing 50% black or brown area on the surface of the peel). Letters A-P represent hybridizations with different cDNA clones
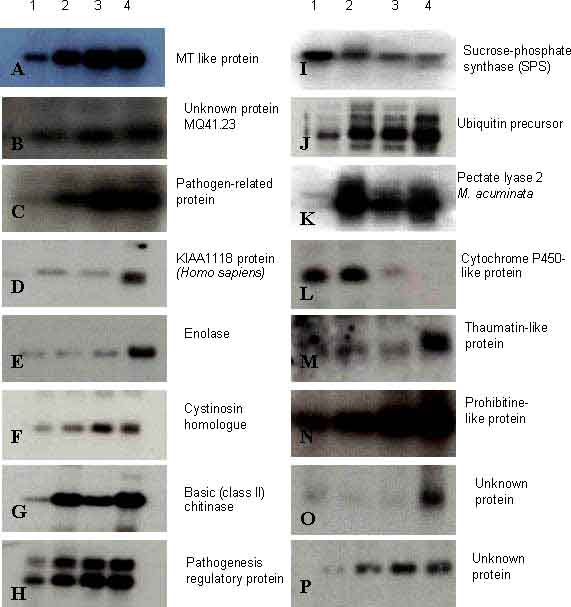
Table 1 Examples of putative annotation of cDNA clones differentially expressed in climacteric and post climacteric stages of banana fruit. Post climacteric fruit were used as tester while pre-climacteric fruit served as driver for the subtraction. SSH libraries were constructed and used as recommended by the manufacturer (Clonetech PCR Select).
|
Access on number |
Size of cDNA Base pairs |
Closest homologue in the NCBI gene-bank |
% Homology Amino acids |
|
gbAAD55980 |
300 |
Extensin-like protein from Zea maize |
82 |
|
gi2497907 |
400 |
Metallothionein-like protein from Musa acuminata |
100 |
|
gi8468019 |
200 |
Putative acetyl transferase from Oryza sativa |
55 |
|
gi11281148 |
500 |
Hypothetical protein T31B5.30 from Arabidopsis thaliana |
78 |
|
gi7485146 |
250 |
Hypothetical protein from Arabidopsis thaliana |
|
|
gi11357666 |
600 |
Hypothetical protein 26G5.140 from Arabidopsis thaliana |
74 |
|
gi13937297 |
300 |
Unknown protein AC087797 from Oryza sativa |
50 |
|
gi34706607 |
250 |
Putative pathogenesis related protein from Oryza sativa |
70 |
|
gi7021736 |
250 |
Putative jasmonic acid regulatory protein from Arabidopsis thaliana |
77 |
|
gi10129675 |
150 |
Beta mannan endo-hydrolase from coffee |
65 |
|
400 |
Pectate lyase from Musa acuminata |
100 |
|
|
400 |
Sucrose phosphate synthase from Musa acuminata |
100 |
|
|
200 |
Cytochrom P450 like protein from Arabidopsis thaliana |
63 |
|
|
gi5042453 |
300 |
Putative pathogenesis related protein from Oryza sativa |
70 |
|
gi82512 |
250 |
Ubiquitin precursor protein from Oryza sativa |
100 |
|
gi2425170 |
300 |
Basic chitinase (class III chitinase) from Oryza sativa |
59 |
|
gi13124057 |
300 |
Cytinosin homologue from Arabidopsis thaliana |
62 |
|
gi134668 |
500 |
Superoxide dismutase precursor from Zea maize |
85 |
|
500 |
No homology |
|
|
|
gi11259780 |
500 |
Protein kinase ANT 1 from Arabidopsis thaliana |
47 |
|
gi11281148 |
250 |
Hypothetical protein T31B5.30 from Arabidopsis thaliana |
60 |
|
gi22273 |
600 |
Enolase from Zea maize |
85 |
5. DEFENSE RELATED GENES FROM THE MUSA GENOME - A COMPARATIVE ANALYSIS
Broadly, resistant genotypes either block the pathogen from entry to susceptible tissues (becomes a non-host) or they exert specific defense mechanisms to localize the invading pathogen. The latter case requires an induced response and involves a signal transduction cascade leading to activation of defense genes. In most cases activation of a defense response requires a recognition event, signaling and finally execution of the defense. Effective defense largely relies on rapid recognition and execution of the protecting mechanism against the pathogen.Perception is mediated by a super-family of genes namely R-genes. Defense mechanisms initiated by R-genes include hypersensitive response (HR) leading to programmed cell death [19-20], synthesis of anti-microbial proteins or metabolites, cell wall reinforcement at the infected site and blockage of vessels [21]. In the majority of cases both resistant and susceptible genotypes share the same downstream defense responses. What differs is the race specific R gene that recognizes the signaling molecule of the pathogen. Susceptible genotypes lack the specific R-gene that perceives the pathogen on the surface of the cell and transmits the signal that activates through a transduction pathway the defense response [22-23]. R-genes in plants are localized in multiple copies in specific loci. Because of promiscuous genetic exchange, paralogs within these clusters exhibit complex evolutionary relationships [24-25]. Strong evidence for high adaptive evolutionary divergence of R-genes is evident from comparative studies. Given the complete sterility of triploid Musa genotypes the likelihood of allelic segregation at a given locus is conceived at a very low frequency. Consequently, adaptive divergence of R-genes as a mechanism to combat pathogens is inferior in these genotypes when compared to their fertile diploid ancestors.
Black and yellow sigatoka (caused by Mycosphaerella fijiensis and Mycosphaerella musicola respectively), have been recognized as the major damaging diseases of bananas almost world-wide [26-27]. Several wild varieties showing resistance or tolerance to the disease have been identified. We are conducting a comparative study of functional genes between a susceptible to a resistant variety using EST analysis combined with selective hybridization libraries. Candidate cDNA clones demonstrating differential expression between the resistant and susceptible genotypes are further analyzed by silencing at the level of mRNA (Figure 3; Table 2).
Figure 3 Northern blot analysis of transcripts from Calcutta 4 and Cavendish leaves probed with clones from Calcutta 4 subtracted from Cavendish cDNA's by SSH technique (as detailed in Table 2). In lanes 1 and 2 (A-D) total RNA (15 µg per lane) from Calcutta 4 leaves was loaded on a 1% agarose gel and blotted onto a nylon membrane while in lanes 3-4 total RNA from Cavendish was loaded
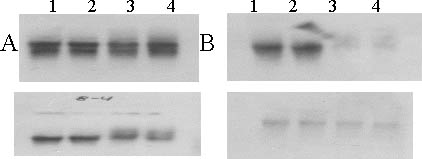
Table 2 Examples of putative annotation of cDNA clones differentially expressed in Calcutta 4 and Cavendish leaves. Calcutta 4 was used as tester while Cavendish cDNA served as driver for the subtraction. SSH libraries were constructed and used as recommended by the manufacturer (Clonetech PCR Select).
|
Accesion number |
Size of cDNA Base pairs |
Closest homologue in the NCBI gene-bank |
% Homology Amino acids |
|
RG-C04-2 |
560 |
resistance gene alike (Aegilops ventricosa) |
68 |
|
RG-B04-3 |
540 |
resistance gene (Avena strigosa) |
87 |
|
RG-A01-7 |
340 |
protein phosphatase 13 (Testis- and skeletal-muscle-specific DSP) (Mus musculus) |
60 |
|
RG-A03-9 |
280 |
OsNAC3 protein (Oryza sativa) |
51 |
|
RG-A08-13 |
465 |
CG2129 gene product (Drosophila melanogaster) |
41 |
|
RG-B04-14 |
440 |
patatin-like protein (Sorghum bicolor) |
83 |
|
RG-D08-5 |
750 |
putative ABC transporter (Arabidopsis thaliana) |
80 |
|
RG-B05-4 |
245 |
putative class I chitinase (Musa acuminata) |
71 |
|
RG- 04-2 |
730 |
Putative protein kinase containing LRR |
77 |
|
RG-14-2 |
200 |
S receptor kinase |
80 |
|
RG-C-1 |
450 |
ADP ribosylation kinase |
78 |
|
RG-C02 |
285 |
Kinesin heavy chain DNA binding protein (Arabidopsis thaliana) |
82 |
|
RG-C04 |
215 |
Putative kinase (Arabidopsis thaliana) |
63 |
|
RG-7-7 |
550 |
Elicitor responsive protein kinase (Arabidopsis thaliana) |
70 |
|
RG-17-3 |
580 |
RAS-related protein RAB7 (Arabidopsis thaliana) |
87 |
|
RG-17-5 |
353 |
Cystein protease component of protease in complex (Oryza sativa). |
59 |
|
RG-7-1 |
260 |
Tryptophane synthase alpha 1- like protein (Arabidopsis thaliana) |
62 |
|
RG-B06 |
425 |
AMP binding protein (Oryzae sativa) |
85 |
REFERENCES
[1] MAY, G. D., et al., Generation of transgenic banana (Musa acuminata) plants via Agrobacterium-mediated transformation, Biotechnology 13 (1995) 486-492.
[2] SAGI, L., et al., Genetic transformation of banana and plantain (Musa spp.) via particle bombardment, Biotechnology 13 (1995) 481-485.
[3] KHAYAT, E., et al., Banana improvement program at Rahan Meristem, Acta Hortic. 490 (1998) 71-78.
[4] BECKER, D. K., et al.,. Genetic transformation of Cavendish banana (Musa spp. AAA group) cv. 'Grand Nain' via microprojectile bombardment, Plant Cell Reports 19 (2000) 229-234.
[5] GANAPATHI, T. R., et al., Agrobacterium-mediated transformation of embryogenic cell suspension of the banana cultivar Rasthali (AAB), Plant Cell Reports 20 (2001) 157-162.
[6] LYSAK, M. A., et al., Flow cytometric analysis of nuclear DNA content in Musa, Theor. Appl. Genet. 98 (1999) 1344-1350.
[7] NORMILE, D., PENNISI, E., Rice: boiled down to bare essentials, Science 296 (2002) 32-36.
[8] McELROY, D., et al., Development of simple transient assay for Ac/Ds activity in cells of intact barley tissue, The Plant J. 11(1) (1997) 157-165.
[9] KAZUHILO, S., et al., Transposition of the maize Ds element from a viral vector to the rice genome, The Plant J. 5(6) (1994) 863-871.
[10] WOLF-EKKEHARD, L., HUIJSER,P., Gene tagging by endogenous transposons, Plant Molecular Biology Manual K1 1-15 (1994) Kluwer Academic Publisher Belgium.
[11] VANCE, V., VAUCHERET, H., et al., RNA silencing in plants - defense and counterdefense, Science 292 (5525) (2001) 2277-2285.
[12] HOFMANN, K., Sensitive protein comparisons with profiles and hidden Markov models, Brief Bioinform. 1(2) (2000) 167-178.
[13] JOHN, P., MARCHAL, J., Ripening and biochemistry of fruit, In: Gowen, S. (Ed.) Banana and Plantains. Chapman and Hall, London (1995) Pp. 434-467.
[14] GIOVANNONI, J., Molecular biology of fruit maturation and ripening, Ann. Rev. of Plant Physiol. Plant Mol. Biology 52 (2001) 725-749.
[15] CLENDENNEN, S. K., MAY, G. D., Differential gene expression in ripening banana fruit, Plant Physiol. 115 (1997) 463-469.
[16] MAUNDER, M., et al., Ethylene stimulates the accumulation of ripening related mRNAs in tomatoes, Plant Cell and Environ, 10 (1987) 177-184.
[17] NAM, Y. W., et al., Isolation and characterization of mRNAs differentially expressed during ripening of wild strawberry (Fragaria vesca L.) fruits, Plant Mol. Biol. 39 (1999) 629-636.
[18] SUYAMA, T., et al., Cloning cDNAs for genes preferentially expressed during fruit growth in cucumber, J. Amer. Soc. Hort. Sci. 124 (1999) 136-139.
[19] GOODMAN, R. M., NOVACK, A. J., et al., The hypersensitivity response of plants to pathogens, A resistance phenomenon, (1994) APS press St. Paul Minnesota.
[20] JONES, A. M., DANGLE, J. I, et al., Logjam at the styx, Programmed cell death in plants, Trends in Plant Sci. 1 (1996) 114-119.
[21] DIXON, R. A., et al., Early events in activation of plant defense response, Ann. Rev. of Plant Phytopath. 32 (1994) 479-501.
[22] GLAZEBROOK, J., Genes controlling expression of defense responses in Arabidopsis - 2001 status, Current Opinion in Plant Biol. 4 (2001) 301-308.
[23] ELLIS, J., et al., Structure, function and evolution of plant disease resistance genes, Current opinions in Plant Biol. 3 (2000) 278-284.
[24] RICHTER, T. E., RONALD, P. C., et al., The evolution of disease resistance, Plant Molecular Biol. 42 (2000) 195-204.
[25] KANAZIN, V., et al., Resistance gene analogs are conserved and clustered in soybean, Proc. Natl. Acad. Sci. USA, 93 (1996) 11746-11750.
[26] MOBAMBO, K. N., et al., Yield loss in plantain from Black Sigatoka leaf spot and field performance of resistant hybrids, Field Crops Res. 35 (1993) 35-42.
[27] BALINT KURTI, P.J., et al., Development of transformation system for Mycosphaerella pathogens of banana: a tool for the study of host pathogen interactions, FEMS Microbiology Letters 195 (2001) 9-15.