Orlando Peixoto de Morais

Orlando Peixoto de Morais
Embrapa Arroz e Feijão, Caixa Postal 179, 75375-000 Santo Antônio
de Goiás, Brazil.
E-mail: [email protected]
Abstract
Today, many plant breeders are extremely concerned about the variability that permits them to obtain, in their projects, significant expressed responses to selection. They know that response to selection is directly proportional to the genetic variance within selection units. However, variation should not be obtained at the cost of reducing the population mean, which would strongly affect selection results. The population mean also constitutes an important factor in reducing the time needed to raise the population to a determined level of mean performance. In this chapter, we discuss the desirability of establishing an equilibrium between means and variances of the traits of the populations developed for improvement through recurrent selection. We also address the advantages of using multiparental crosses, common in the creation of such populations, as a way of initially increasing allelic frequency and guaranteeing the presence of favourable alleles in all loci of interest.
Resumen
Hoy en día existe entre los fitomejoradores una extrema preocupación por la variabilidad que permita obtener respuestas significativas en sus trabajos. Se sabe que la respuesta a la selección es directamente proporcional a la varianza genética entre las unidades de selección. Sin embargo, no se debe conseguir variación a expensas de la reducción de la media de la población, pues esta afecta fuertemente el resultado del proceso de selección y constituye un factor importante en la reducción del tiempo necesario para elevar la población a un determinado nivel de desempeño medio. En este capítulo, se discutirá la conveniencia del establecimiento de un equilibrio entre media y varianza de las características de las poblaciones desarrolladas, para ser mejoradas por medio del método de selección recurrente. Además se abordarán las ventajas de los cruzamientos multiparentales, comunes en la creación de esas poblaciones, como medio de aumentar inicialmente la frecuencia alélica y garantizar la presencia de alelos favorables en todos los loci de interés.
Plant breeders recognize the great difficulty of selecting for quantitative traits, particularly for complex traits such as yield. Such traits are affected by the simultaneous action of various genes, usually located on different chromosomes. Even so, the use of conventional methods of genetic improvement has permitted significant progress for such traits. However, in rice, increasing difficulties have recently been hindering this progress, as described below.
To better illustrate this aspect, a trait that segregates for only 20 loci will be assumed. Under the most favourable conditions, the frequency of individuals in an endogamous population that carries all the desired alleles would be a little more than 9.5 × 10-7. This would correspond to the selection of only two alleles with intermediate frequency (equal to 0.5) per locus. For this case, a population of 1:9.5 × 10-7 would have to be evaluated, that is, more than 1 million individuals would be needed to identify at least one individual with favourable alleles in all 20 loci. In reality, a much smaller sample can be used to identify one of these individuals, depending on the required probability of success. However, if this probability is very high, a sample of even more than 1 million individuals may have to be evaluated.
The example discussed has a binomial distribution. The n number of individuals for evaluation can be estimated so that at least one individual carrying favourable alleles in the 20 loci can be identified at whatever level of probability. For example, for a probability level P at 99%, n would equal 4 828 869 individuals. This number would become 7 243 303, should P have a value of 99.9%.
The same would occur for a level as low as P = 1%. The number of individuals to be sampled and evaluated would be 10 539. This would still be very high, considering the difficulties of evaluating the trait in question such as experiments with replications and preferably being carried out at more than one site.
For all these reasons, an alternative is needed to increase yield, in rice varieties. One is to seek achieving the selection of one or more individuals with favourable alleles in all 20 loci, not all at once, but by stages. This permits thinking about selecting a group of individuals with an average performance that is better than that of the population base from which it was derived. These individuals can be used as progenitors of a new population that will present an average allelic frequency that is more favourable than that of the first population. The increase in allelic frequency also increases the possibility of selecting individuals with a larger number of favourable alleles (Table 1). This cyclic and systematic process of selecting desirable individuals in a population, followed by a recombination of the same to form a new population with a higher potential than the first for the intended goals, is called recurrent selection (Fehr, 1987).
Table 1. Frequency of individuals with favourable alleles in all loci in a population segregating for 20 loci, under three conditions of allelic frequency.
|
Generation |
Allelic frequency (p) |
||
|
p = ¾ |
p = ½ |
p = ¼ |
|
|
S0 |
1:3.6 |
1:315 |
1:3.6 × 107 |
|
S1 |
1:30.0 |
1:12 089 |
1:1.9 × 109 |
|
S2 |
1:94.0 |
1:99 437 |
1:3.5 × 1010 |
|
... |
... |
... |
... |
|
... |
... |
... |
... |
|
... |
... |
... |
... |
|
S8 |
1:315.0 |
1:1 048 576 |
1:1.1 × 1012 |
However, where the same population is used in its highest heterotic state (S0), the frequency of individuals with favourable alleles changes from 1 in 1 048 576 to only 1 in 315 individuals (Morais, 2001). This means that the frequency of the genotype under selection will increase by almost 3328 times.
Moreover, the use of S0 individuals or of families derived from them (families S0:n, with n preferably being small) presents two additional advantages: a shorter selection cycle and higher selection pressure, as the number of selection units in terms of selecting pure lines can be reduced by half. All this is achieved without altering the effective size of the selected sample (Morais, 1997).
Ramalho (1997) very convincingly presents the advantages of adopting recurrent selection. He includes for the case where N homozygous lines are extracted by crossing the two best lines originating from a first selection cycle. (These are thus second-cycle lines.) For this case, and assuming a trait influenced by 40 independent loci, Ramalho adapts Fouilloux and Bannerot (1988) to obtain, in only one selection cycle, the same cumulative result of two selection cycles. He found the resources used must be several tens of times more (Table 2). For a hypothetical case of complete heritability and evaluating 200 individuals, to obtain in one cycle the total of results obtained in two, then the evaluated population must be 290 times larger. That is, 116 000 individuals must be evaluated, and certainly with equally higher expenditures of resources.
Selection is always effective through changes in allelic frequency that, in practice, are not tangible. Together with the evolving selection process, the population’s observable properties, such as means, variances and covariances of the principal traits, should be evaluated during successive selection cycles. These evaluations must be implemented at the same point of the individuals’ life cycles, as much for the sampled population’s age as for its genetic structure.
Table 2. Considering a trait influenced by 40 loci, the numbers of lines needed to obtain a given number of favourable alleles are compared between one case of two selection cycles and another case of one.
|
Two cyclesa |
One cycleb |
Ratio N/n |
|
|
n |
Lfa |
N |
|
|
50 |
31.3 |
3 500 |
35 |
|
100 |
32.6 |
18 800 |
94 |
|
200 |
33.8 |
116 000 |
290 |
|
400 |
34.8 |
543 200 |
679 |
a. n refers to number of individuals evaluated by selection cycle, in the case of two cycles; Lfa refers to expected number of loci with favourable alleles in the best-performing line from the second cycle.
b. N refers to number of lines to be evaluated in only one selection cycle so that the best-performing line has the same Lfa as its counterpart in the second cycle, in the case of two selection cycles.
Source: Ramalho (1997).
Falconer (1987), in his detailed discussion, shows that response to selection cannot be expected to occur indefinitely. Sooner or later, all the favourable alleles that were originally segregating will become fixed and, at this point, the population will no longer respond to selection.
Because of their desire to obtain significant responses, plant breeders are highly concerned with variation. The search for that variation should not, despite everything, bring negative consequences to the magnitude of the initial values of the means of the traits of interest in the populations being selected.
This chapter discusses the desirability of establishing an equilibrium between means and variances of the traits of populations developed for improvement through recurrent selection.
As Chaves (1997) suggests, the mean of population Xn, after n selection cycles, can be obtained through the following formula: n
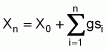
where:
X0 is the estimate of the mean of the population base before the first selection cycle
gsi is the gain due to selection in cycle i, when i = 1, 2, 3,..., n
The initial mean of the population base (X0) can clearly be seen to directly affect the result of selection. It constitutes an important factor in reducing the time needed to raise a population to a given level of mean performance. Based on this, during the creation of a population for recurrent selection, all available alternatives must be used so that its initial mean will be high.
Because of the similarity of the creative processes, for populations of autogamous species destined for recurrent selection, equations of prediction of averages developed to estimate averages of synthetic varieties can be used.
To serve as the basis for a recurrent selection programme, a population of an autogamous species created from pure lines, combined two by two, presents, when in S0 (maximum heterozygosity), traits similar to the synthetic varieties of allogamous species. Hence, as Chaves (1997) suggests, the population’s average performance can be predicted from information on its k progenitors and its ‘k(k - 1)/2’ hybrid combinations, using the following formula of Equation 1:
where:
Gj corresponds to the average of progenitor j
Cjj' is the average of crosses between progenitors j and j'
As a result, to decide which progenitors should be effectively involved in the composition of a population, we need to obtain information from a diallelic cross that encompasses all the potential progenitors and their biparental crosses. According to Vencovsky and Barriga (1992), the number of possible populations that can be synthesized from k progenitors results from the formula 2k-(k+1), each with a predictable mean based on information from the diallelic cross, involving k progenitors.
In equation 1, the population mean is seen as a function of the average of progenitors and their hybrid combinations. Thus, the progenitors chosen should not only have good performance per se, but each should also have a high capacity for combination with the other progenitors. That is, each should be divergent with respect to the set of selected progenitors.
While executing a project to exploit genetic resources, using recurrent selection, the evaluation units are normally families of known ancestry but with a high degree of heterozygosity. Hence, recombination of selected evaluation units constitutes a process identical with that of forming composites of allogamous species, whose means can be predicted by using an equation similar to equation 1. Thus, the progenitors (G) may be families selected in the previous cycle and Cjj' represents the biparental crosses between the selected families j and j'.
It should be noted that, when the mixture of seeds of families possessing the male-sterility gene is used as a recombination procedure, as mentioned by Châtel and Guimarães (1995), all descendants have a structure similar to that which would be obtained with the mixture of all seeds from a diallelic cross, which should not alter the population mean.
The contribution of genetic variation to the efficiency of the selection process must be analysed. Response to selection is directly proportional to the genetic variance among the selection units, but that increase in genetic variation should not be sought by reducing the population mean. Greater genetic variance must be found by using divergent progenitors, which should be identified from elite progenitors that perform the best for the traits of interest. The divergent elite progenitors not only condition greater genetic variability, but they also present, among themselves, a higher capacity for combination, providing a population with a higher general mean, as shown above.
Below, we discuss the importance of using divergent progenitors and of raised performance in biparental crosses. We then present the relative advantages of using multiparental populations.
Biparental populations
To better illustrate this point, we will assume the following:
By crossing two divergent progenitors, we will analyse the efficiency of selection in two different cases:
Case 1: Presence of favourable alleles in 80% of the loci in one parent and in 20% in the other parent, with maximum divergence, that is, the hybrid population segregates for all 20 loci
Case 2: Both parents possess favourable alleles in 80% of the loci but also express maximum divergence, that is, the hybrid population segregates for 8 loci (0.2×20×2=8)
Considering that, for each locus, the genotypic value designated for homozygotes equals the unit (a = 1), we will evaluate the following parameters for the two cases:
Mean
Using the classic formula:
m = m + a(p - q) + 2dpq
(Falconer 1987) and considering µ = 100, the mean of the population segregating for all loci (Case 1), with a frequency p = q = 0.5, where a = 1 and d = 0 (genotypic value of the heterozygote), is:
For parents (homozygotes) with favourable alleles in 80% of the loci (16 loci with favourable alleles and 4 with unfavourable alleles):
m = 100 + 1(1 - 0)16 + 1(0 - 1)4 = 112
For parents with favourable alleles in 20% of the loci:
m = 100 + 1(1 - 0)4 + 1(0 - 1)16 = 88
Hybrid populations
Case 1 (segregating for all 20 loci, with an intermediate allelic frequency):
m = 100 + 1(0.5 - 0.5)20 = 100
Case 2 (alleles fixed in 12 loci and segregating for 8 loci, with an intermediate allelic frequency):
m = 100 + 1(1 - 0)12 + 1(0.5 - 0.5)8 = 112
Range of variation (R)
This is the difference between the genotypic values of those individuals with favourable alleles fixed in all 20 loci and the values of those individuals with no favourable alleles in the segregating hybrid population:
Case 1 (n = 20):
R = 2na = 2 × 20 × 1 = 40
Case 2 (n = 8):
R = 2na = 2 × 8 × 1 = 16
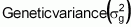
For one locus, without dominance, it is:
Considering n loci, with identical a values, not linked and no epistatic interaction, the equation for genetic variance becomes:
Then, the estimate can be finalized as follows:
Case 1:
Case 2:
The population originating only from elite progenitors (Case
2) presents less variability, that is, both its range of variation (R) and its
genetic variance 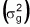 correspond
to 40% of the values present in Case 1. However, its mean is almost 12% greater.
Consequently, the difference between the means of the two populations
corresponds to 60% of the total response to possible selection, using the first
population. That is, more than half of the gain that could possibly be obtained
with the Case 1 population can be achieved by simply using the Case 2
population, which is synthesized with elite progenitors.
correspond
to 40% of the values present in Case 1. However, its mean is almost 12% greater.
Consequently, the difference between the means of the two populations
corresponds to 60% of the total response to possible selection, using the first
population. That is, more than half of the gain that could possibly be obtained
with the Case 1 population can be achieved by simply using the Case 2
population, which is synthesized with elite progenitors.
Another way of evaluating the current value of the two populations is to estimate, for the two populations, the t number of individuals that should be sampled, so that, at a given level of probability (e.g. 99%), at least one will carry favourable alleles in all loci. In the Case 1 population, the frequency of individuals with favourable alleles (in homozygosis or heterozygosis) at all 20 loci is obtained by p1=(3/4)20. Consequently, (1-p1) represents the frequency of individuals with favourable alleles in less than 20 loci.
When t individuals are sampled from this population, the probability that all will have fewer than 20 loci with favourable alleles is expressed as [1-(3/4)20]t. It suffices to estimate whether the value of t at this probability will be equal to or less than 0.01 by using logarithms:
To evaluate at least 1450 individuals for a trait that is influenced by 20 loci and is strongly subjected to environmental action will certainly require the adoption of strong environmental control and replication. The evaluation of the first population, seeking to identify at least one individual with favourable alleles in all 20 loci, would therefore be both difficult and wasteful. However, for the second population (Case 2), this task would be entirely viable because, on conducting similar calculations, the conclusion is that the minimum number of individuals to be evaluated is only 44.
To continue comparing the two populations for improvement, the performance of both can still be analysed when they are submitted to recurrent selection, a process of selection that can induce, for each locus, changes in gene frequency (Table 3).
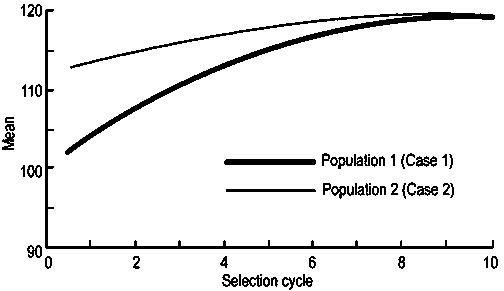
In this case, after 10 selection cycles, all the alleles would be fixed by the 10th cycle, when the populations present low levels of genetic variability and almost the same means. Table 3 shows that Case 1 still presents a genetic variance that is more than twice that of Case 2, and that its responses to selection are much more expressive. This is exactly what enraptures plant breeders: high response to selection.
In practice, however, Case 2 presents, overall, a higher potential for selection of superior lines because, as well as presenting genetic variability that can be governed by the same objective as for Case 1, it always shows, in intermediate phases, a higher mean and greater capacity to offer superior lines. The superiority of Case 2 is particularly notable in the initial cycles, as con- firmed by comparing the estimates of t, where t is the number of individuals S0 or of S0:n families that should be evaluated, so that there is a 99% certainty of having at least one individual that carries favourable alleles in all loci.
As selection cycles are conducted, differences between the two populations are reduced, but the synthesized population based only on the elite progenitors (Case 2) is invariably always superior, as clearly shown in Figure 1.
In any population under evaluation, the following questions must be asked:
How many of its individuals or families derived from self-pollination should be evaluated so that, admitting an error of less than 1%, at least one will surpass the population mean by two standard deviations?
How many loci with favourable alleles would this individual be expected to have for the two populations under study?
Table 3. Allelic frequency (p), alteration of p as a function
of selection 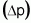 , population
means in successive selection cycles, responses to selection (Rs), genetic
variance
, population
means in successive selection cycles, responses to selection (Rs), genetic
variance 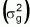 and estimates of t
and I1 for biparental populations synthesized by involving one elite
progenitor and one non-elite (Case 1), and only elite progenitors (Case
2).
and estimates of t
and I1 for biparental populations synthesized by involving one elite
progenitor and one non-elite (Case 1), and only elite progenitors (Case
2).
|
Generation |
p
|
Dp |
Mean |
Rs |
|
t |
I1 |
|||||
|
1 |
2 |
1 |
2 |
1 |
2 |
1 |
2 |
1 |
2 |
|||
|
0 |
0.500 |
|
100.0 |
112.0 |
|
|
10.0000 |
4.0000 |
1450 |
44 |
20.0 |
21.2 |
|
1 |
0.600 |
0.100 |
100.4 |
113.6 |
4.0 |
1.6 |
9.6000 |
3.8400 |
148 |
16 |
21.0 |
21.4 |
|
2 |
0.690 |
0.090 |
107.6 |
115.0 |
3.6 |
1.4 |
8.5560 |
3.4224 |
32 |
8 |
21.5 |
21.4 |
|
3 |
0.770 |
0.080 |
110.8 |
116.3 |
3.2 |
1.3 |
7.0840 |
2.8336 |
11 |
4 |
21.5 |
21.2 |
|
4 |
0.830 |
0.060 |
113.2 |
117.3 |
2.4 |
1.0 |
5.6440 |
2.2576 |
6 |
3 |
21.4 |
21.0 |
|
5 |
0.880 |
0.050 |
115.2 |
118.1 |
2.0 |
0.8 |
4.2240 |
1.6896 |
3 |
2 |
21.1 |
20.8 |
|
6 |
0.920 |
0.040 |
116.8 |
118.7 |
1.6 |
0.6 |
2.9440 |
1.1776 |
2 |
2 |
20.8 |
20.5 |
|
7 |
0.950 |
0.030 |
118.0 |
119.2 |
1.2 |
0.5 |
1.9000 |
0.7600 |
2 |
1 |
20.5 |
20.3 |
|
8 |
0.970 |
0.020 |
118.8 |
119.5 |
0.8 |
0.3 |
1.1640 |
0.4656 |
1 |
1 |
20.3 |
20.2 |
|
9 |
0.980 |
0.010 |
119.2 |
119.7 |
0.4 |
0.2 |
0.7840 |
0.3136 |
1 |
1 |
20.2 |
20.1 |
|
10 |
0.985 |
0.005 |
119.4 |
119.8 |
0.2 |
0.1 |
0.5910 |
0.2364 |
1 |
1 |
20.2 |
20.1 |
To answer the first question, the assumption is made that the trait being evaluated follows a normal distribution- a common assumption in population analyses. This distribution has about 2.27% of individuals situated to the right of the point corresponding to the mean (m) and at more than two standard deviations (s), that is, m+2s. Individuals of a population can thus be separated into two classes: one that is situated to the right of m+2s at a probability of P1=0.0227, and the other to the left of this same point of reference at a probability of 1-P1=0.9773. With these two classes of individuals, the binomial distribution can be used again. Thus, when r individuals of a population are sampled, the possibilities available are such as listed in Table 4.
All the r first possibilities serve the desired situation; only the last does not. It suffices then, to estimate the value of r that brings the (r + 1)th probability to less than 0.01, that is:
Figure 1. Evolution of the means of two case populations over 10 recurrent selection cycles.
Because 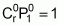 ,
whatever the value of r and P 1, the following is obtained:
,
whatever the value of r and P 1, the following is obtained:
(1-P1)r £ 0.01 [2]
Again, applying logarithms to equation 2, the following is obtained:
As a result, if more than 201 individuals or families of that population derived from self-pollination are evaluated, at least one will surpass the population mean by two or more standard deviations 99% of times this evaluation is carried out.
Now, to answer the second question, we need to estimate the probable number of loci with favourable alleles that a best-performing individual of each population studied should have, evaluating a sample of 201 individuals.
The only loci to be considered are those in segregation (n = 20 in the first population [Case 1] and n = 8 in the second [Case 2]), with an allelic frequency of p that is identical for all these n loci. In this case, the number of loci with favourable alleles among individuals follows a binomial distribution with a mean of m=np2 and variance =np2(1-p2).
Considering each locus separately, the value of p2 is given as p2=1-(1-p)2, with p being the values described in Table 3. When n is large, the binomial distribution appears normal. In this situation, the number of loci with favourable alleles in an individual can be estimated by the formula li=m+c. Value "c" is the value of the abscissa corresponding to that individual’s position in a descending classification of all the individuals in the sample in terms of the number of loci with favourable alleles (Morais, 2001).
Table 4. The number of individuals situated to the right of a reference point in a normal distribution and the respective probabilities of occurrence.
|
Possibility |
Number of individuals to the right of a reference point |
Probability |
|
1 |
r individuals (all) |
|
|
2 |
r - 1 individuals |
|
|
3 |
r - 2 individuals |
|
|
4 |
r - 3 individuals |
|
|
|
... |
... |
|
|
... |
... |
|
|
... |
... |
|
r - 1 |
2 individuals |
|
|
R |
1 individual |
|
|
r + 1 |
0 (no) individuals |
|
Thus, for a population with a sample of 201 individuals, the value of c corresponding to the first individual of a greater number of favourable alleles is given by the value of the abscissa at the point of truncation for a selection percentile w = 1/201 = 0.00497. The value of c can be obtained by exploring tables of referents of the normal distribution pattern that exist in statistical compendia such as that of Pimentel Gomes (1978). With this procedure, the value obtained was c = 2.58.
All the values of l1, estimated by the equation above for the two populations (Cases 1 and 2), are found in the last two columns of Table 3. For Case 1, the value of l1 was expected to be 20, as the value of r is only 201, that is, much less than the value of t, where p = 0.5, in the same table.
The differences of the estimated values for l1 probably originate from considering the binomial distribution as a normal distribution for values of n as low as 8 and 20. For example, attributing the value of 100 to n and considering p = 0.6 (p2 = 0.84), then the values of t and l1 are estimated to be 257 874 460.2 and 93, respectively. For 100 loci, segregating at a frequency for favourable alleles of p = 0.6, identical for all loci, then a sample of almost 258 million must be taken-more than the Brazilian population-so that, at a 99% probability, at least one individual with favourable alleles in all 100 loci would be included.
In contrast, in the case of the sample of 201 individuals, the best performing for the trait under consideration should carry favourable alleles in 93 loci, admitting the same frequencies of error.
Multiparental populations
So far, we have considered a population resulting from a biparental cross. What would be the advantages of involving several progenitors in the synthesis of a population? Normally, one would imagine that, with several progenitors, greater variation is obtained. This is not always true. For any number of segregating loci, two contrasting progenitors can be identified with respect to these same loci. Thus, greater variation would be obtained because the allelic frequencies for the segregating loci would, invariably, be intermediate. Before listing the advantages of multiparental crosses, we will analyse some of the characteristics of their genetic constitution.
Whether assuming the same trait considered for the biparental populations just analysed or conditioned for the expression of genes of 20 loci, without epistatic interaction of any kind, then this case could involve, within its constitution, as many as C16 20 = 4845 progenitors with favourable alleles in 16 of the 20 loci available and be equally divergent in relation to a larger number of loci. All these progenitors would present similar means (112, in this case), for the trait under consideration.
When analysing
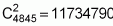 possible progenitors, each
would present the same characteristics as the biparental population Case 2, that
is, with an intermediate allelic frequency, a mean equal to 112 and all the
other derived characteristics.
possible progenitors, each
would present the same characteristics as the biparental population Case 2, that
is, with an intermediate allelic frequency, a mean equal to 112 and all the
other derived characteristics.
Considered as a whole, this hypothetical multiparental population would also present a mean equal to 112, but its initial allelic frequency would be spectacularly superior.
This demonstrates that a special population such as this one, made up of n homozygous progenitors, each with favourable alleles in s loci, would present an initial allelic frequency as follows:
Replacing n and s by 20 and 16, respectively, we obtain p = 0.80.
To involve a larger number of progenitors would be even more advantageous, but the number of segregating loci would increase in tandem, instead of having some fixed with favourable alleles, as was observed for Case 2, composed of only two divergent elite progenitors.
Selection would already be initiated in a population base of high value, saving time and resources to achieve the final objective. The population, from the beginning of the selection process, becomes promising as a source of lines that, in autogamous species, represent a commercially usable form. On average, one of each 8 individuals of population S0 would carry favourable alleles in all 20 loci.
Where this population is subjected to self-pollination until complete homozygosis, the frequency of best-performing plants, with all the 20 desired alleles in homozygosis, would be 1 in 397 and, as such, still easily identified by using adequate experimental designs.
The population’s performance in a selection process would be similar to that of Case 2 (Table 3) from a phase that is intermediate between cycles 3 and 4.
To conclude, two important aspects are emphasized for those who work with population improvement, not only in rice, but also in any other crop. They are:
In genetic improvement, those procedures that increase the genetic variability of the populations being studied should be used, but they should not lead to reduced means in the traits of interest
The use of a large number of elite and genetically divergent progenitors will help acquire a larger initial allelic frequency and a higher probability of having a population base with favourable alleles in all loci of interest.
References
Châtel, M. & Guimarães, E.P. 1995. Selección recurrente con androesterilidad en arroz. Cali, Colombia, CIRAD-CA & CIAT. 70 pp.
Chaves, L.J. 1997. Criterios para escoger proprogenitores para programa de selección recurrente. In E.P. Guimarães, ed. Selección recurrente en arroz, pp. 13-24. Cali, Colombia, CIAT.
Falconer, D.S. 1987. Introdução à genética quantitativa. Transl. by Martinho de Alemida e Silva and José Carlos Silva. Viçosa, MG, Brazil, Universidade Federal de Viçosa (UFV). 279 pp.
Fehr, W.R. 1987. Principles of cultivar development: theory and technique. Vol. 1. New York, Macmillan Publishing. 536 pp.
Fouilloux, G. & Bannerot, H. 1988. Selection methods in the common bean (Phaseolus vulgaris L.). In P. Gepts, ed. Genetics resources of Phaseolus beans, pp. 503-542. Dordrecht, Netherlands, Kluwer.
Morais, O.P. de. 1997. Tamaño efectivo de la población. In E.P. Guimarães, ed. Selección recurrente en arroz, pp. 25-44. Cali, Colombia, CIAT.
Morais, O.P. de. 2001. Seleção recorrente em aotógamas. In 1º Congresso Brasileiro de Melhoramento de Plantas, Goiânia-GO, 3-5 April 2001. Sociedade Brasileira de Melhoramento de Plantas. (Available at www.sbmp.org.br).
Pimentel Gomes, F. 1978. Iniciação à estatística. São Paulo, Brazil, Editora Nobel. 211 pp.
Ramalho, M.A.P. 1997. Melhoramento do Feijoeiro. In A. de F.B. Abreu; F.M.A. Gonçalves; O.G. Marques, Jr & P.H.E. Ribeiro, eds. Proc. Simpósio sobre atualização en genética e melhoramento de plantas, pp. 167-196. Lavras, MG, Brazil, Universidade Federal de Lavras (UFLA).
Vencovsky, R. & Barriga, P. 1992. Genética biométrica no fitomelhoramento. Ribeirão Preto, Brazil, Revista Brasileira de Genética. 496 pp.