


Table 3.10. Values of pH and organic carbon content observed in soil samples collected from natural forest.
|
Soil pit |
pH (x) |
Organic carbon (%) (y) |
(x2) |
(y2) |
(xy) |
|
1 |
5.7 |
2.10 |
32.49 |
4.4100 |
11.97 |
|
2 |
6.1 |
2.17 |
37.21 |
4.7089 |
13.24 |
|
3 |
5.2 |
1.97 |
27.04 |
3.8809 |
10.24 |
|
4 |
5.7 |
1.39 |
32.49 |
1.9321 |
7.92 |
|
5 |
5.6 |
2.26 |
31.36 |
5.1076 |
12.66 |
|
6 |
5.1 |
1.29 |
26.01 |
1.6641 |
6.58 |
|
7 |
5.8 |
1.17 |
33.64 |
1.3689 |
6.79 |
|
8 |
5.5 |
1.14 |
30.25 |
1.2996 |
6.27 |
|
9 |
5.4 |
2.09 |
29.16 |
4.3681 |
11.29 |
|
10 |
5.9 |
1.01 |
34.81 |
1.0201 |
5.96 |
|
11 |
5.3 |
0.89 |
28.09 |
0.7921 |
4.72 |
|
12 |
5.4 |
1.60 |
29.16 |
2.5600 |
8.64 |
|
13 |
5.1 |
0.90 |
26.01 |
0.8100 |
4.59 |
|
14 |
5.1 |
1.01 |
26.01 |
1.0201 |
5.15 |
|
15 |
5.2 |
1.21 |
27.04 |
1.4641 |
6.29 |
|
Total |
82.1 |
22.2 |
450.77 |
36.4100 |
122.30 |
The steps to be followed in the computation of correlation coefficient are as follows.
Step 1. Compute covariance of x and y and variances of both x and y using Equation (3.24).
Cov (x,y) = ![]()
= 0.05
V(x) = 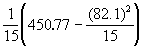
= 0.0940
V(y) = 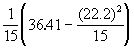
= 0.2367
Step 2. Compute the correlation coefficient using Equation (3.24).
r = ![]()
= 0.3541
3.8.1. Testing the significance of correlation coefficient.
A value of correlation coefficient obtained from a sample needs to be tested for significance to confirm if a real relationship exists between the two variables in the population considered. It is usual to set up the null hypothesis as ![]() against the alternative hypothesis,
against the alternative hypothesis, ![]() .
.
For relatively small n, the null hypothesis that ![]() can be tested using the test statistic,
can be tested using the test statistic,
![]() (3.25)
(3.25)
This test statistic is distributed as Student’s t with n-2 degrees of freedom.
Consider the data given in Table 3.10 for which n = 15 and r = 0.3541. To test as ![]() against
against ![]() , we compute the test statistic using Equation (3.25).
, we compute the test statistic using Equation (3.25).
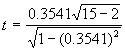 = 1.3652
= 1.3652
Referring Appendix 2, the critical value of t is 2.160, for 13 degrees of freedom at the probability level, a = 0.05. Since the computed t value is less than the critical value, we conclude that the pH and organic carbon content measured from soil samples are not significantly correlated. For convenience one may use Appendix 5 which gives values of correlation coefficients beyond which an observed correlation coefficient can be declared as significant for a certain number of observations at a desired level of significance.
In order to test the hypothesis that ![]() where r
0 is any specified value of r
, Fisher’s z-transformation is employed which is given by,
where r
0 is any specified value of r
, Fisher’s z-transformation is employed which is given by,
![]() (3.26)
(3.26)
where ln indicates natural logarithm.
For testing the null hypothesis, we use the test statistic,
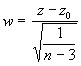 (3.27)
(3.27)
where 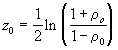
The statistic w follows a standard normal distribution.
For the purpose of illustration, consider the data given in Table 3.10 for which n = 15 and r = 0.3541. Suppose that we have to test ![]() = 0.6. For testing this null hypothesis, the values of r and r
are first subjected to z transformation.
= 0.6. For testing this null hypothesis, the values of r and r
are first subjected to z transformation.
![]() = 0.3701
= 0.3701
![]() = 0.6932
= 0.6932
The value of the test statistic would be,
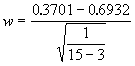 = 1.16495
= 1.16495
Since the value of w is less than 1.96, the critical value, it is nonsignificant at 5% level of significance. Hence we may conclude that the correlation coefficient between pH and organic carbon content in the population is not significantly different from 0.6.
Correlation coefficient measures the extent of interrelation between two variables which are simultaneously changing with mutually extended effects. In certain cases, changes in one variable are brought about by changes in a related variable but there need not be any mutual dependence. In other words, one variable is considered to be dependent on the other variable changes, in which are governed by extraneous factors. Relationship between variables of this kind is known as regression. When such relationships are expressed mathematically, it will enable us to predict the value of one variable from the knowledge of the other. For instance, the photosynthetic and transpiration rates of trees are found to depend on atmospheric conditions like temperature or humidity but it is unusual to expect a reverse relationship. However, in many cases, it so happens that the declaration of certain variables as independent is made only in a statistical sense although when reverse effects are conceivable in such cases. For instance, in a volume prediction equation, tree volume is taken to be dependent on dbh although the dbh cannot be considered as independent of the effects of tree volume in a physical sense. For this reason, independent variables in the context of regression are sometimes referred as regressor variables and the dependent variable is called the regressand.
The dependent variable is usually denoted by y and the independent variable by x. When only two variables are involved in regression, the functional relationship is known as simple regression. If the relationship between the two variables is linear, it is known as simple linear regression, otherwise it is known as nonlinear regression. When one variable is dependent on two or more independent variables, the functional relationship between the dependent and the set of independent variables is known as multiple regression. For the sake of easiness in description, only the case of simple linear regression is considered here. Reference is made to Montgomery and Peck (1982) for more complex cases.
3.9.1. Simple linear regression
The simple linear regression of y on x in the population is expressible as
![]() (3.28)
(3.28)
where ![]() and
and ![]() are parameters also known as regression coefficients and e
is a random deviation possible from the expected relation. But for e
with a mean value of zero, Equation (3.28) represents a straight line with a
as the intercept and b
as the slope of the line. In other words, a
is the expected value of y when x assumes the value zero and b
gives the expected change in y for a unit change in x. The slope of a linear regression line may be positive, negative or zero depending on the relation between y and x.
are parameters also known as regression coefficients and e
is a random deviation possible from the expected relation. But for e
with a mean value of zero, Equation (3.28) represents a straight line with a
as the intercept and b
as the slope of the line. In other words, a
is the expected value of y when x assumes the value zero and b
gives the expected change in y for a unit change in x. The slope of a linear regression line may be positive, negative or zero depending on the relation between y and x.
In practical applications, the values of a and b are to be estimated from observations made on y and x variables from a sample. For instance, to estimate the parameters of a regression equation proposed between atmospheric temperature and transpiration rate of trees, a number of paired observations are made on transpiration rate and temperature at different times of the day from a number of trees. Let such pairs of values be designated as (xi, yi); i = 1, 2, . . ., n where n is the number of independent pairs of observations. The values of a and b are estimated using the method of least squares (Montgomery and Peck, 1982) such that the sum of squares of the difference between the observed and expected value is minimum. In the estimation process, the following assumptions are made viz., (i) The x values are non-random or fixed (ii) For any given x, the variance of y is the same (iii) The y values observed at different values of x are completely independent. Appropriate changes will need to be made in the analysis when some of these assumptions are not met by the data. For the purpose of testing hypothesis of parameters, an additional assumption of normality of errors will be required.
In effect, the values of a and b are obtained from the formulae,
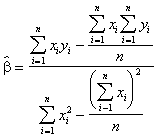 (3.29)
(3.29)
![]() (3.30)
(3.30)
The equation ![]() is taken as the fitted regression line which can be used to predict the average value of the dependent variable, y, associated with a particular value of the independent variable, x. Generally, it is safer to restrict such predictions within the range of x values in the data.
is taken as the fitted regression line which can be used to predict the average value of the dependent variable, y, associated with a particular value of the independent variable, x. Generally, it is safer to restrict such predictions within the range of x values in the data.
The standard errors of ![]() can be estimated by the following formulae.
can be estimated by the following formulae.
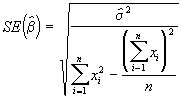 (3.31)
(3.31)
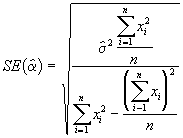 (3.32)
(3.32)
where 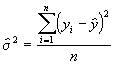
The standard error of an estimate is the standard deviation of the sampling distribution of that estimate and is indicative of the extent of reliability of that estimate.
As an example, consider the data presented in Table 3.11 which contain paired values of photosynthetic rate and light interception observed on leaves of a particular tree species. In this example, the dependent variable is photosynthetic rate and the independent variable is the quantity of light. The computations involved in fitting a regression line are given in the following.
Step 1. Compute the values of numerator and denominator of Equation (3.29) using the sums, sum of squares and sum of products of x and y generated in Table 3.5.
![]() =
= ![]() = 2.6906
= 2.6906
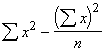 = 12.70 -
= 12.70 - ![]() = 0.1508
= 0.1508
Table 3.11. Data on photosynthetic rate in ![]() mol m-2s-1 (y) along with the measurement of radiation in mol m-2s-1 (x) observed on a tree species.
mol m-2s-1 (y) along with the measurement of radiation in mol m-2s-1 (x) observed on a tree species.
|
X |
y |
x2 |
xy |
|
0.7619 |
7.58 |
0.58 |
5.78 |
|
0.7684 |
9.46 |
0.59 |
7.27 |
|
0.7961 |
10.76 |
0.63 |
8.57 |
|
0.8380 |
11.51 |
0.70 |
9.65 |
|
0.8381 |
11.68 |
0.70 |
9.79 |
|
0.8435 |
12.68 |
0.71 |
10.70 |
|
0.8599 |
12.76 |
0.74 |
10.97 |
|
0.9209 |
13.73 |
0.85 |
12.64 |
|
0.9993 |
13.89 |
1.00 |
13.88 |
|
1.0041 |
13.97 |
1.01 |
14.02 |
|
1.0089 |
14.05 |
1.02 |
14.17 |
|
1.0137 |
14.13 |
1.03 |
14.32 |
|
1.0184 |
14.20 |
1.04 |
14.47 |
|
1.0232 |
14.28 |
1.05 |
14.62 |
|
1.0280 |
14.36 |
1.06 |
14.77 |
|
|
|
|
|
Step 2. Compute the estimate of a and b using Equations (3.29) and (3.30).
![]() = 17.8422
= 17.8422
![]() 12.60 - (17.8421)(0.9148)
12.60 - (17.8421)(0.9148)
= -3.7202
The fitted regression line is ![]() which can be used to predict the value of photosynthetic rate at any particular level of radiation within the range of data. Thus, the expected photosynthetic rate at 1 mol m-2s-1 of light would be,
which can be used to predict the value of photosynthetic rate at any particular level of radiation within the range of data. Thus, the expected photosynthetic rate at 1 mol m-2s-1 of light would be,
![]() = 14.122
= 14.122
Step 3. Get an estimate of s 2 as defined in Equation (3.32).
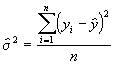 = 0.6966
= 0.6966
Step 4. Develop estimates of standard errors of ![]() using Equations (3.31) and (3.32).
using Equations (3.31) and (3.32).
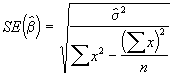 =
=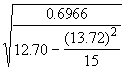 = 2.1495
= 2.1495
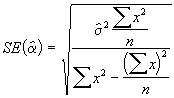 =
= 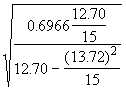 = 1.9778
= 1.9778
3.9.2. Testing the significance of the regression coefficient
Once the regression function parameters have been estimated, the next step in regression analysis is to test the statistical significance of the regression function. It is usual to set the null hypothesis as H0: b = 0 against the alternative hypothesis, H1: b ¹ 0 or (H1: b < 0 or H1: b > 0, depending on the anticipated nature of relation). For the testing, we may use the analysis of variance procedure. The concept of analysis of variance has already been explained in Section 3.6 but its application in the context of regression is shown below using the data given in Table 3.11.
Step1. Construct an outline of analysis of variance table as follows.
Table 3.12. Schematic representation of analysis of variance for regression analysis.
|
Source of variation |
Degree of freedom (df) |
Sum of squares (SS) |
Mean square
|
Computed F |
|
Due to regression |
1 |
SSR |
MSR |
|
|
Deviation from regression |
n-2 |
SSE |
MSE |
|
|
Total |
n-1 |
SSTO |
Step 2. Compute the different sums of squares as follows.
Total sum of squares = 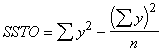 (3.33)
(3.33)
= (7.58)2 + (9.46)2 + . . . + (14.36)2 - ![]()
= 58.3514
Sum of square due to regression = SSR = 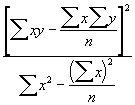 (3.34)
(3.34)
= ![]()
= 48.0062
Sum of squares due to deviation from regression = SSE = SSTO - SSR (3.35)
=58.3514 - 48.0062 = 10.3452
Step 3. Enter the values of sums of squares in the analysis of variance table as in Table 3.13 and perform the rest of the calculations.
Table 3.13. Analysis of variance for the regression equation derived for data in Table 3.11.
|
Source of variation |
Degree of freedom (df) |
Sum of squares (SS) |
Mean square
|
Computed F at 5% |
|
Due to regression |
1 |
48.0062 |
48.0062 |
60.3244 |
|
Deviation from regression |
13 |
10.3452 |
0.7958 |
|
|
Total |
14 |
58.3514 |
Step 4.Compare the computed value of F with tabular value at (1,n-2) degrees of freedom. For our example, the calculated value (60.3244) is greater than the tabular value of F of 4.67 at (1,13) degrees of freedom at 5% level of significance and so the F value is significant. If the computed F value is significant, then we can state that the regression coefficient, b is significantly different from 0. The sum of squares due to regression when expressed as a proportion of the total sum of squares is known as the coefficient of determination which measures the amount of variation in y accounted by the variation in x. In other words, coefficient of determination indicates the proportion of the variation in the dependent variable explained by the model. For the present example, the coefficient of determination (R2) is
![]() (3.36)
(3.36)
![]()
= 0.8255
In analysis of variance, generally, the significance of any known component of variation is assessed in comparison to the unexplained residual variation. Hence, proper control is to be exercised to reduce the magnitude of the uncontrolled variation. Either the model is expanded by including more known sources of variation or deliberate control is made on many variables affecting the response. Otherwise, any genuine group differences would go undetected in the presence of the large residual variation. In many instances, the initial variation existing among the observational units is largely responsible for the variation in their further responses and it becomes necessary to eliminate the influence of inherent variation among the subjects from the comparison of the groups under consideration. Analysis of covariance is one of the methods used for the purpose of reducing the magnitude of unexplained error. For instance, in an experimental context, covariance analysis can be applied when observations on one or more correlated variables are available from each of the experimental units along with the observations on the response variable under study. These additional related variables are called covariates or ancillary or concomitant variables. It is necessary that these variables are associated with the variable under study. For example, in yield trials, variation in the initial stocking induced by extraneous factors, residual effects of the previous crops grown in the site etc., can serve as covariates.
Analysis of covariance is a synthesis of the methods of the analysis of variance and those of regression. The concept is elaborated here in the context of an experiment with just one variable under study denoted by y and a single covariate denoted by x. Let there be t experimental groups to be compared, each group consisting of r experimental units. The underlying model in this case could be
![]() (3.37)
(3.37)
where yij is the response observed on the jth experimental unit belonging to ith group,
(i = 1, 2, …, t; j = 1, 2, …, r)
m y is the overall population mean of y,
a i is the effect of being in the ith group,
b is the within group regression coefficient of y on x
xij is the observation on ancillary variate on jth unit of ith group.
m x is the overall mean of the covariate
eij ‘s are the error components which are assumed to be normally and independently distributed with zero mean and a constant variance ![]() .
.
Covariance analysis is essentially an extension of the analysis of variance and hence, all the assumptions for a valid analysis of variance apply here as well. In addition, the covariance analysis requires that (i) The relationship between the primary character of interest y and the covariate x is linear (ii) The strength of the relation between y and x remains the same in each experimental group (iii) The variation in the covariate should not have been a result of the group differences.
The steps involved in the analysis of covariance are explained below.
Step 1. The first step in the analysis of covariance is to compute the sum of squares due to the different components, for the variate y and the covariate x in the usual manner of analysis of variance. The computation formulae for the same are given below.
Total SS of y = SSTO(y) = 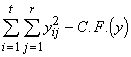 (3.38)
(3.38)
where 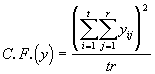 (3.39)
(3.39)
Group SS of y = SSG(y) = 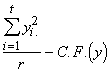 (3.40)
(3.40)
Error SS of y = SSE(y) = SSTO(y) -SSG(y) (3.41)
Total SS of x = SSTO(x) = 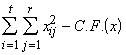 (3.42)
(3.42)
where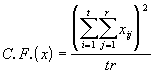 (3.43)
(3.43)
Group SS of x = SSG(x) = 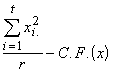 (3.44)
(3.44)
Error SS of x = SSE(x) = SSTO(x) - SSG(x) (3.45)
Step 2. Compute the sum of products for x and y as follows.
Total SP = SPTO(xy) = 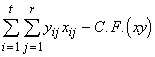 (3.46)
(3.46)
where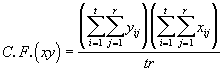 (3.47)
(3.47)
Group SP = SPG(xy) = 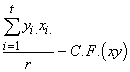 (3.48)
(3.48)
Error SP = SPE(xy) = SSTO(xy) - SSG(xy) (3.49)
Step 3. The next step is to verify whether the covariate is affected by the experimental groups. If x is not affected by the groups, there should not be significant differences between groups with respect to x. The regression co-efficient within groups is computed as
![]() (3.50)
(3.50)
The significance of ![]() is tested using F-test. The test statistic F is given by
is tested using F-test. The test statistic F is given by
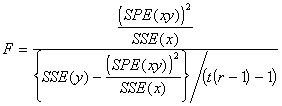 (3.51)
(3.51)
The F statistic follows a F distribution with 1 and t(r-1)-1 degrees of freedom. If the regression coefficient is significant, we proceed to make adjustments in the sum of squares for y for the variation in x. If not significant, it is not worthwhile to make the adjustments.
Step 4. Adjusted values for y are computed as follows:
Adjusted total SS of y = Adj. SSTO(y) = SSTO(y) - 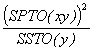 (3.52)
(3.52)
Adjusted error SS of y = Adj. SSE(y) = SSE(y) -  (3.53)
(3.53)
Adjusted group SS of y = Adj. SSG(y)= Adj. SSTO(y) - Adj. SSE(y) (3.54)
Conventionally, the above results are combined in a single table as in Table 3.14.
Step 5. The adjusted group means are obtained by the formula,
![]() (3.55)
(3.55)
The standard error for the difference between two adjusted means is given by :
SE(d) = 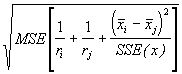 (3.56)
(3.56)
where the symbols have the usual meanings.
When the number of replications is the same for all the groups and when averaged over all values of ![]() we get,
we get,
SE(d) = 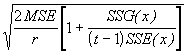 (3.57)
(3.57)
Table 3.14. Analysis of covariance (ANOCOVA) table
|
Source of variation |
df |
Sum of squares and products |
||
|
y |
x |
xy |
||
|
Total |
tr-1 |
SSTO(y) |
SSTO(x) |
SPTO(xy) |
|
Group |
t-1 |
SSG(y) |
SSG(x) |
SPG(xy) |
|
Error |
t(r-1) |
SSE(y) |
SSE(x) |
SPE(xy) |
Table 3.14. Cond…
|
Adjusted values for y |
||||
|
Source of variation |
df |
SS |
MS |
F |
|
Total |
tr-2 |
Adj. SSTO(y) |
- |
- |
|
Group |
- |
- |
- |
- |
|
Error |
t(r-1)-1 |
Adj. SSE(y) |
MSE |
- |
|
Adj. Group |
t-1 |
Adj. SSG(y) |
MSG |
|
Let us use the data given in Table 3.15 to demonstrate how the analysis of covariance is carried out. The data represent plot averages based on forty observations of initial height (x) and height attained after four months (y) of three varieties of ipil ipil, (Leucaena leucocephala), each grown in 10 plots in an experimental site.
Table 3.15. Initial height (x) and height after four months (y) in cm of three varieties of ipil ipil (Leucaena leucocephala), in an experimental area.
|
Plot |
Variety 1 |
Variety 2 |
Variety 3 |
|||
|
x |
y |
x |
y |
x |
y |
|
|
1 |
18 |
145 |
27 |
161 |
31 |
180 |
|
2 |
22 |
149 |
28 |
164 |
27 |
158 |
|
3 |
26 |
156 |
27 |
172 |
34 |
183 |
|
4 |
19 |
151 |
25 |
160 |
32 |
175 |
|
5 |
15 |
143 |
21 |
166 |
35 |
195 |
|
6 |
25 |
152 |
30 |
175 |
36 |
196 |
|
7 |
16 |
144 |
21 |
156 |
35 |
187 |
|
8 |
28 |
154 |
30 |
175 |
23 |
137 |
|
9 |
23 |
150 |
22 |
158 |
34 |
184 |
|
10 |
24 |
151 |
25 |
165 |
32 |
184 |
|
Total |
216 |
1495 |
256 |
1652 |
319 |
1789 |
|
Mean |
21.6 |
149.5 |
25.6 |
165.2 |
31.2 |
178.9 |
The analysis is carried out following the format shown in Table 3.14. The computations are demonstrated below:
Step 1.Compute sum of squares for x and y variables using Equations (3.38) to (3.45).
C.F.(y) = ![]() = 812136.5333
= 812136.5333
SSTO(y) = (145)2 + (149)2 + . . . + (184)2 - 812136.5333
= 7493.4667
SSG(y) = ![]()
= 4328.4667
SSE(y) = 7493.4667 - 4328.4667
= 3615.0
C.F.(x) = ![]()
= 20856.0333
SSTO(x) = (18)2 + (22)2 + . . . + (32)2 -20.856.0333
= 966.9697
SSG(x) = ![]()
= 539.267
SSE(x) = 966.9697-539.267
= 427.7027
Step 2.Compute sum of products for x and y variables using Equations (3.46) to (3.49).
![]()
![]()
= 130145.8667
SPTO(xy) = 18(145) + 22(149) + . . . +32(184) -130145.8667
= 2407.1333
SPG(xy) = ![]()
= 1506.44
SPE(xy) = 2407.1333 - 1506.44 = 900.6933
Step 3. Compute the regression coefficient and test its significance using Equations (3.50) and (3.51).
![]() =
= ![]()
= 2.1059
The significance of ![]() is tested using F-test. The test statistic F is given by the Equation (3.51).
is tested using F-test. The test statistic F is given by the Equation (3.51).
F = 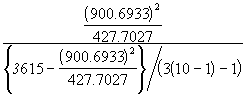
= ![]()
= 28.7012
Table value of F with (1,26) degrees of freedom = 9.41 at 5% level of significance. Here calculated value of F is grater than tabular value and hence ![]() is significantly different from zero.
is significantly different from zero.
Step 4. Compute adjusted sums of squares for the different sources in the ANOCOVA using Equations (3.52) to (3.54). Summarise the results as in Table 3.14 and compute mean square values for group (MSG) and error (MSE) and also the value of F based on these mean squares.
Adj. SSTO(y) = ![]()
= 1501.2513
Adj. SSE(y) = ![]()
= 1268.2422
Adj. SSG(y) = 1501.2513 - 1268.2422
= 233.0091
MSG = ![]() = 116.5046
= 116.5046
MSE = ![]()
= 48.7785
![]()
= ![]()
= 2.39
Table 3.16. Analysis of covariance table for the data in Table 3.15.
|
Sources of variation |
df |
Sum of squares and products |
Adjusted values for y |
|||||
|
y |
x |
xy |
df |
SS |
MS |
F |
||
|
Total |
29 |
7493.467 |
966.970 |
2407.133 |
28 |
1501.25 |
- |
- |
|
Group |
2 |
4328.467 |
539.267 |
1506.440 |
- |
- |
- |
- |
|
Error |
27 |
3615.000 |
427.703 |
900.693 |
26 |
1268.24 |
48.8 |
- |
|
Group adjusted for the covariate |
2 |
233.009 |
116.5 |
2.4 |
||||
The value of F for (2,26) degrees of freedom at 5% level of significance is 3.37. Since the observed F value, 2.4, is less than the critical value, we conclude that there are no significant differences among the varieties.
Step 5. Get the adjusted group means and standard error of the difference between any two adjusted group means by using Equations (3.55) and (3.57).
![]() = 149.5 - 2.1059(21.6 - 26.37) = 159.54
= 149.5 - 2.1059(21.6 - 26.37) = 159.54
![]() = 165.2 - 2.1059(25.6 - 26.37) = 166.82
= 165.2 - 2.1059(25.6 - 26.37) = 166.82
![]() = 178.9 - 2.1059(31.2 - 26.37) = 168.73
= 178.9 - 2.1059(31.2 - 26.37) = 168.73
SE(d) = 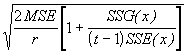
= 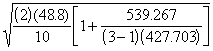 = 3.9891
= 3.9891
The standard error of the difference between group means will be useful in pairwise comparison of group means as explained in Chapter 4.
3.11 Analysis of repeated measures
Repeated measurements of observational units are very frequent in forestry research. The term repeated is used to describe measurements which are made of the same characteristic on the same observational unit but on more than one occasion. In longitudinal studies, individuals may be monitored over a period of time to record the changes occurring in their states. Typical examples are periodical measurements on diameter or height of trees in a silvicultural trial, observations on disease progress on a set of seedlings in a nursery trial, etc. Repeated measures may be spatial rather than temporal. For instance, consider measurements on wood characteristics of several stems at the bottom, middle and top portion of each stem and each set of stems coming from a different species. Another example would be that of soil properties observed from multiple core samples at 0-15, 15-50 and 50-100 cm depth from different types of vegetation.
The distinctive feature of repeated measurements is the possibility of correlations between successive measurements over space or time. Autocorrelation among the residuals arising on account of repeated measurements on the same experimental units leads to violation of the basic assumption of independence of errors for conducting an ordinary analysis of variance. However, several different ways of analysing repeated measurements are available. These methods vary in their efficiency and appropriateness depending upon the nature of data. If the variance of errors at each of the successive measurements is the same and also the covariances between errors of different measurement occasions are the same, one may choose to subject the data to a ‘univariate mixed model analysis’. In case the errors are unstructured, a multivariate analysis is suggestible taking repeated measurements as different characters observed on the same entities (Crowder and Hand, 1990). The details of univariate analysis are illustrated below under a simplified observational set up whereas the reader is referred to (Crowder and Hand, 1990) for multivariate analysis in this context.
The general layout here is that of n individuals x p occasions with the individuals divided into t groups of size ni (i = 1, 2, …, t). Let the hypothesis to be tested involve a comparison among the groups. The model used is
yijk = m + a i+ eij + b j + g ij + eijk (3.58)
where yijk is the observation on kth individual in the ith group at jth occasion;
( i =1, …, t, j =1, …, p, k =1, …, ni.)
m is the general mean,
a i is the effect of ith level of the factor ‘group’,
b j is the effect of jth level of the factor ‘occasion’,
g ij is the interaction effect for the ith level of the factor ‘group’ and jth level of the factor ‘occasion’. This term measures the differences between the groups with respect to their pattern of response over occasions. More elaborate discussion on interaction is included in Chapter 4.
In model (3.58), the random component eij are assumed to be independently and normally distributed with mean zero and variance![]() and eijk is the random error component which is also assumed to be independently and normally distributed with mean zero and variance
and eijk is the random error component which is also assumed to be independently and normally distributed with mean zero and variance ![]() . In the model, a
i ’s and b
j’s are assumed to be fixed.
. In the model, a
i ’s and b
j’s are assumed to be fixed.
Let yi.. denote the total of all observations under the ith level of factor, group; y.j. denote the total of all observations under the jth level of factor occasion; yij. denote the total of all observations in the (ij)th cell; y… denote the grand total of all the observations. These notations are expressed mathematically as
yi.. = ![]() , y.j. =
, y.j. = ![]() , yij. =
, yij. = ![]() , y… =
, y… = ![]()
The univariate mixed model ANOVA is shown below.
Table 3.17. Schematic representation of univariate mixed model analysis of variance.
|
Sources of variation |
Degrees of freedom |
Sum of squares |
Mean sum of squares |
F-ratio |
|
Group |
t-1 |
|
|
|
|
Individuals within groups |
|
|
|
|
|
Occasion |
p-1 |
|
|
|
|
Occasion x Group |
(t-1)(p-1) |
|
|
|
|
Occasion x Individuals within groups |
|
|
|
|
|
Total |
|
SSTO |
The computational formulae for the sum of squares in the above table are as follows,
SSTO = ![]()
![]() (3.59)
(3.59)
SSG =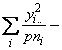
![]() (3.60)
(3.60)
SSEa = 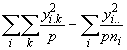 (3.61)
(3.61)
SSO =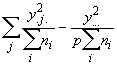 (3.62)
(3.62)
SSOG = 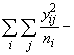
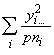
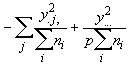 (3.63)
(3.63)
SSEb = SST - SSG - SSEa - SSO - SSOG (3.64)
For illustration of the analysis, consider the data given in Table 3.18. The data represent the mycelial growth (mm) of five isolates of Rizoctonia solani on PDA medium after 14, 22, 30 and 38 hours of incubation, each isolate grown in three units of the medium. Here, the isolates represent ‘groups’ and different time points represent the ‘occasions’ of Table 3.17.
Table 3.18. Data on mycelial growth (mm) of five groups of R. solani isolates on PDA medium.
|
Mycelial growth (mm) observed at different occasions |
|||||
|
R. Solani isolate |
PDA unit |
14 hr. |
22 hr. |
30 hr. |
38 hr. |
|
1 |
1 |
29.00 |
41.00 |
55.00 |
68.50 |
|
2 |
28.00 |
40.00 |
54.00 |
68.50 |
|
|
3 |
29.00 |
42.00 |
55.00 |
69.00 |
|
|
2 |
1 |
33.50 |
46.50 |
59.00 |
74.00 |
|
2 |
31.50 |
44.50 |
58.00 |
71.50 |
|
|
3 |
29.00 |
42.50 |
56.50 |
69.00 |
|
|
3 |
1 |
26.50 |
38.00 |
48.50 |
59.50 |
|
2 |
30.00 |
40.00 |
50.00 |
61.00 |
|
|
3 |
26.50 |
38.00 |
49.50 |
61.00 |
|
|
4 |
1 |
48.50 |
67.50 |
75.50 |
83.50 |
|
2 |
46.50 |
62.50 |
73.50 |
83.50 |
|
|
3 |
49.00 |
65.00 |
73.50 |
83.50 |
|
|
5 |
1 |
34.00 |
41.00 |
51.00 |
61.00 |
|
2 |
34.50 |
44.50 |
55.50 |
67.00 |
|
|
3 |
31.00 |
43.00 |
53.50 |
64.00 |
|
|
Total |
506.50 |
696.00 |
868.00 |
1044.50 |
|
Analysis of the above data can be conducted as follows.
Step 1. Compute the total sum of squares using Equation (3.59) with values of Table 3.18.
SSTO = ![]()
![]()
Step 2.Construct an Isolate x PDA unit two-way table of totals by summing up the observations over different occasions and compute the marginal totals as shown in Table 3.19. Compute SSG and SSEa using the values in this table and Equations (3.60) and (3.61).
Table 3.19. The Isolate x PDA unit totals computed from data in Table 3.18.
|
Isolates |
||||||
|
PDA unit |
1 |
2 |
3 |
4 |
5 |
Total |
|
1 |
193.50 |
213.00 |
172.50 |
275.00 |
187.00 |
1041.00 |
|
2 |
190.50 |
205.50 |
181.00 |
266.00 |
201.50 |
1044.50 |
|
3 |
195.00 |
197.00 |
175.00 |
271.00 |
191.50 |
1029.50 |
|
Total |
579.00 |
615.50 |
528.50 |
812.00 |
580.00 |
3115.00 |
SSG =![]()
![]()
= 4041.04
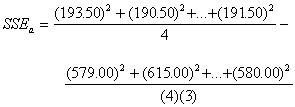
= 81.92
Step 3. Form the Isolate x Occasion two-way table of totals and compute the marginal totals as shown in Table 3.20. Compute SSO, SSOG and SSEb using Equations (3.62) to (3.64).
Table 3.20. The Isolate x Occasion table of totals computed from data in Table 3.18
|
Occasion |
|||||
|
Isolate |
14 hr. |
22 hr. |
30 hr. |
38 hr. |
Total |
|
1 |
86.00 |
123.00 |
164.00 |
206.00 |
579.00 |
|
2 |
94.00 |
133.50 |
173.50 |
214.50 |
615.50 |
|
3 |
83.00 |
116.00 |
148.00 |
181.50 |
528.50 |
|
4 |
144.00 |
195.00 |
222.50 |
250.50 |
812.00 |
|
5 |
99.50 |
128.50 |
160.00 |
192.00 |
580.00 |
|
Total |
506.50 |
696.00 |
868.00 |
1044.50 |
3115.00 |
SSO =![]()
= 10637.08
SSOG =![]()
![]()
![]()
= 172.46
SSEb = ![]()
= 29.08
Step 4. Summarise the results as in Table 3.21 and perform the remaining calculations to obtain the mean squares and F-ratios using the equations reported in Table 3.17.
Table 3.21. ANOVA table for the data in Table 3.18.
|
Sources of variation |
Degrees of freedom |
Sum of squares |
Mean sum of squares |
F-ratio |
|
Group |
4 |
4041.04 |
1010.26 |
123.33* |
|
Individuals within groups |
10 |
81.92 |
8.19 |
|
|
Occasion |
3 |
10637.08 |
3545.69 |
3657.45* |
|
Occasion x Group |
12 |
172.46 |
14.37 |
14.82* |
|
Occasion x Individuals within groups |
30 |
29.08 |
0.97 |
|
|
Total |
59 |
14961.58 |
Compare the computed values of F with tabular values of F with corresponding degrees of freedom at the desired probability level. All the computed F values in the above table are greater than corresponding tabular F values. Hence, we conclude that the variation due to groups, occasion and their interaction are significant meaning essentially that the isolates differ in their growth pattern across time.