


4. DESIGN AND ANALYSIS OF EXPERIMENTS
Planning an experiment to obtain appropriate data and drawing inference out of the data with respect to any problem under investigation is known as design and analysis of experiments. This might range anywhere from the formulations of the objectives of the experiment in clear terms to the final stage of the drafting reports incorporating the important findings of the enquiry. The structuring of the dependent and independent variables, the choice of their levels in the experiment, the type of experimental material to be used, the method of the manipulation of the variables on the experimental material, the method of recording and tabulation of data, the mode of analysis of the material, the method of drawing sound and valid inference etc. are all intermediary details that go with the design and analysis of an experiment.
4.1 Principles of experimentation
Almost all experiments involve the three basic principles, viz., randomization, replication and local control. These three principles are, in a way, complementary to each other in trying to increase the accuracy of the experiment and to provide a valid test of significance, retaining at the same time the distinctive features of their roles in any experiment. Before we actually go into the details of these three principles, it would be useful to understand certain generic terms in the theory experimental designs and also understand the nature of variation among observations in an experiment.
Before conducting an experiment, an experimental unit is to be defined. For example, a leaf, a tree or a collection of adjacent trees may be an experimental unit. An experimental unit is also sometimes referred as plot. A collection of plots is termed a block. Observations made on experimental units vary considerably. These variations are partly produced by the manipulation of certain variables of interest generally called treatments, built-in and manipulated deliberately in the experiment to study their influences. For instance, clones in clonal trials, levels and kinds of fertilizers in fertilizer trials etc. can be called treatments. Besides the variations produced in the observations due to these known sources, the variations are also produced by a large number of unknown sources such as uncontrolled variation in extraneous factors related to the environment, genetic variations in the experimental material other than that due to treatments, etc. They are there, unavoidable and inherent in the very process of experimentation. These variations because of their undesirable influences are called experimental error thereby meaning not an arithmetical error but variations produced by a set of unknown factors beyond the control of the experimenter.
It is further interesting to note that these errors introduced into the experimental observations by extraneous factors may be either systematic or random in their mode of incidence. The errors arising due to an equipment like a spring balance which goes out of calibration due to continued use or the error due to observerís fatigue are examples of systematic error. On the other hand, the unpredictable variation in the amount of leaves collected in litter traps under a particular treatment in a related experiment is random in nature. It is clear that any number of repeated measurements would not overcome systematic error where as it is very likely that the random errors would cancel out with repeated measurements. The three basic principle viz., randomization, replication and local control are devices to avoid the systematic error and to control the random error.
4.1.1. Randomization
Assigning the treatments or factors to be tested to the experimental units according to definite laws or probability is technically known as randomization. It is the randomization in its strict technical sense, that guarantees the elimination of systematic error. It further ensures that whatever error component that still persists in the observations is purely random in nature. This provides a basis for making a valid estimate of random fluctuations which is so essential in testing of significance of genuine differences.
Through randomization, every experimental unit will have the same chance of receiving any treatment. If, for instance, there are five clones of eucalyptus to be tried in say 25 plots, randomization ensures that certain clones will not be favoured or handicapped by extraneous sources of variation over which the experimenter has no control or over which he chooses not to exercise his control. The process of random allocation may be done in several ways, either by drawing lots or by drawing numbers from a page of random numbers, the page itself being selected at random. The method is illustrated in later sections dealing with individual forms of experimental designs.
Replication
Replication is the repetition of experiment under identical conditions but in the context of experimental designs, it refers to the number of distinct experimental units under the same treatment. Replication, with randomization, will provide a basis for estimating the error variance. In the absence of randomization, any amount of replication may not lead to a true estimate of error. The greater the number of replications, greater is the precision in the experiment.
The number of replications to be included in any experiment depends upon many factors like the homogeneity of experimental material, the number of treatments, the degree of precision required etc. As a rough rule, it may be stated that the number of replications in a design should provide at least 10 to 15 degrees of freedom for computing the experimental error variance.
4.1.3. Local control
Local control means the control of all factors except the ones about which we are investigating. Local control, like replication is yet another device to reduce or control the variation due to extraneous factors and increase the precision of the experiment. If, for instance, an experimental field is heterogeneous with respect of soil fertility, then the field can be divided into smaller blocks such that plots within each block tend to be more homogeneous. This kind of homogeneity of plots (experiment units) ensures an unbiased comparison of treatment means, as otherwise it would be difficult to attribute the mean difference between two treatments solely to differences between treatments when the plot differences also persist. This type of local control to achieve homogeneity of experimental units, will not only increase the accuracy of the experiment, but also help in arriving at valid conclusions.
In short, it may be mentioned that while randomization is a method of eliminating a systematic error (i.e., bias) in allocation thereby leaving only random error component of variation, the other two viz., replication and local control try to keep this random error as low as possible. All the three however are essential for making a valid estimate of error variance and to provide a valid test of significance.
4.2. Completely randomized design
A completely randomized design (CRD) is one where the treatments are assigned completely at random so that each experimental unit has the same chance of receiving any one treatment. For the CRD, any difference among experimental units receiving the same treatment is considered as experimental error. Hence, CRD is appropriate only for experiments with homogeneous experimental units, such as laboratory experiments, where environmental effects are relatively easy to control. For field experiments, where there is generally large variation among experimental plots in such environmental factors as soil, the CRD is rarely used.
4.2.1. Layout
The step-by-step procedure for randomization and layout of a CRD are given here for a pot culture experiment with four treatments A, B, C and D, each replicated five times.
Step 1. Determine the total number of experimental plots (n) as the product of the number of treatments (t) and the number of replications (r); that is, n = rt. For our example, n = 5 x 4 = 20. Here, one pot with a single plant in it may be called a plot. In case the number of replications is not the same for all the treatments, the total number of experimental pots is to be obtained as the sum of the replications for each treatment. i.e.,
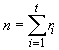 where ri is the number of times the ith treatment replicated
where ri is the number of times the ith treatment replicated
Step 2. Assign a plot number to each experimental plot in any convenient manner; for example, consecutively from 1 to n.
Step 3. Assign the treatments to the experimental plots randomly using a table of random numbers as follows. Locate a starting point in a table of random numbers (Appendix 6) by closing your eyes and pointing a finger to any position in a page. For our example, the starting point is taken at the intersection of the sixth row and the twelfth (single) column of two-digit numbers. Using the starting point obtained, read downward vertically to obtain n = 20 distinct two-digit random numbers. For our example, starting at the intersection of the sixth row and the twelfth column, the 20 distinct two-digit random numbers are as shown here together with their corresponding sequence of appearance.
Random number : 37, 80, 76, 02, 65, 27, 54, 77, 48, 73,
Sequence : 1, 2, 3, 4, 5, 6, 7, 8, 9, 10,
Random number : 86, 30, 67, 05, 50, 31, 04, 18, 41, 89
Sequence : 11, 12, 13, 14, 15, 16, 17, 18, 19, 20
Rank the n random numbers obtained in ascending or descending order. For our example, the 20 random numbers are ranked from the smallest to the largest, as shown in the following:
|
Random Number |
Sequence |
Rank |
Random Number |
Sequence |
Rank |
|
37 |
1 |
8 |
86 |
11 |
19 |
|
80 |
2 |
18 |
30 |
12 |
6 |
|
76 |
3 |
16 |
67 |
13 |
14 |
|
02 |
4 |
1 |
05 |
14 |
3 |
|
65 |
5 |
13 |
50 |
15 |
11 |
|
27 |
6 |
5 |
31 |
16 |
7 |
|
54 |
7 |
12 |
04 |
17 |
2 |
|
77 |
8 |
17 |
18 |
18 |
4 |
|
48 |
9 |
10 |
41 |
19 |
9 |
|
73 |
10 |
15 |
89 |
20 |
20 |
Divide the n ranks derived into t groups, each consisting of r numbers, according to the sequence in which the random numbers appeared. For our example, the 20 ranks are divided into four groups, each consisting of five numbers, as follows:
|
Group Number |
Ranks in the Group |
||||
|
1 |
8 |
13 |
10 |
14 |
2 |
|
2 |
18 |
5 |
15 |
3 |
4 |
|
3 |
16 |
12 |
19 |
11 |
9 |
|
4 |
1 |
17 |
6 |
7 |
20 |
Assign the t treatments to the n experimental plots, by using the group number as the treatment number and the corresponding ranks in each group as the plot number in which the corresponding treatment is to be assigned. For our example, the first group is assigned to treatment A and plots numbered 8, 13, 10, 14 and 2 are assigned to receive this treatment; the second group is assigned to treatment B with plots numbered 18, 5, 15, 3 and 4; the third group is assigned to treatment C with plots numbered 16, 12, 19, 11 and 9; and the fourth group to treatment D with plots numbered 1, 17, 6, 7 and 20. The final layout of the experiment is shown Figure 4.1.
|
Plot no Treatment |
1 D |
2 A |
3 B |
4 B |
|
5 B |
6 D |
7 D |
8 A |
|
|
9 C |
10 A |
11 C |
12 C |
|
|
13 A |
14 A |
15 B |
16 C |
|
|
17 D |
18 B |
19 C |
20 D |
Figure 4.1. A sample layout of a completely randomised design with four treatments (A, B, C and D) each replicated five times
4.2.2. Analysis of variance
There are two sources of variation among the n observations obtained from a CRD trial. One is the variation due to treatments, the other is experimental error. The relative size of the two is used to indicate whether the observed difference among treatments is real or is due to chance. The treatment difference is said to be real if treatment variation is sufficiently larger than experimental error.
A major advantage of the CRD is the simplicity in the computation of its analysis of variance, especially when the number of replications is not uniform for all treatments. For most other designs, the analysis of variance becomes complicated when the loss of data in some plots results in unequal replications among the treatments tested.
The steps involved in the analysis of variance for data from a CRD experiment with unequal number of replications are given below. The formulae are easily adaptable to the case of equal replications and hence not shown separately. For illustration, data from a laboratory experiment are used, in which observations were made on mycelial growth of different Rizoctonia solani isolates on PDA medium (Table 4.1).
Step 1. Group the data by treatments and calculate the treatment totals (Ti) and grand total (G). For our example, the results are shown in Table 4.1 itself.
Step 2. Construct an outline of ANOVA table as in Table 4.2.
Table 4.1. Mycelial growth in terms of diameter of the colony (mm) of R. solani isolates on PDA medium after 14 hours of incubation.
|
R. solani isolates |
Mycelial growth |
Treatment total |
Treatment mean |
||
|
Repl. 1 |
Repl. 2 |
Repl. 3 |
(Ti) |
||
|
RS 1 |
29.0 |
28.0 |
29.0 |
86.0 |
28.67 |
|
RS 2 |
33.5 |
31.5 |
29.0 |
94.0 |
31.33 |
|
RS 3 |
26.5 |
30.0 |
56.5 |
28.25 |
|
|
RS 4 |
48.5 |
46.5 |
49.0 |
144.0 |
48.00 |
|
RS 5 |
34.5 |
31.0 |
65.5 |
32.72 |
|
|
Grand total |
446.0 |
||||
|
Grand mean |
34.31 |
||||
Table 4.2. Schematic representation of ANOVA of CRD with unequal replications.
|
Source of variation |
Degree of freedom (df) |
Sum of squares (SS) |
Mean square
|
Computed F |
|
Treatment |
t - 1 |
SST |
MST |
|
|
Error |
n - t |
SSE |
MSE |
|
|
Total |
n - 1 |
SSTO |
Step 3.With the treatment totals (Ti) and the grand total (G) of Table 4.1, compute the correction factor and the various sums of squares, as follows. Let yij represent the observation on the jth PDA medium belonging to the ith isolate; i = 1, 2, Ö, t ; j = 1, 2, Ö, ri..
![]() (4.1)
(4.1)
![]()
= 15301.23
![]() (4.2)
(4.2)
=![]()
= 789.27
SST = 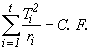 (4.3)
(4.3)
= 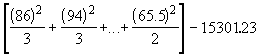
= 762.69
SSE = SSTO - SST (4.4)
= 789.27 - 762.69 = 26.58
Step 4. Enter all the values of sums of squares in the ANOVA table and compute the mean squares and F value as shown in the Table 4.2.
Step 5. Obtain the tabular F values from Appendix 3, with f1 and f2 degrees of freedom where f1 = treatment df = (t - 1) and f2 = error df = (n - t), respectively. For our example, the tabular F value with f1 = 4 and f2 = 8 degrees of freedom is 3.84 at 5% level of significance. The above results are shown in Table 4.3.
Table 4.3. ANOVA of mycelial growth data of Table 4.1.
|
Source of variation |
Degree of freedom |
Sum of squares |
Mean square |
Computed F |
Tabular F 5% |
|
Treatment |
4 |
762.69 |
190.67 |
57.38* |
3.84 |
|
Error |
8 |
26.58 |
3.32 |
||
|
Total |
12 |
789.27 |
* Significant at 5% level
Step 7. Compare the computed F value of Step 4 with the tabular F value of Step 5, and decide on the significance of the difference among treatments using the following rules:
(i) If the computed F value is larger than the tabular F value at 5% level of significance, the variation due to treatments is said to be significant. Such a result is generally indicated by placing an asterisk on the computed F value in the analysis of variance.
(ii) If the computed F value is smaller than or equal to the tabular F value at 5% level of significance, the variation due to treatments is said to be nonsignificant. Such a result is indicated by placing ns on the computed F value in the analysis of variance or by leaving the F value without any such marking.
Note that a nonsignificant F in the analysis of variance indicates the failure of the experiment to detect any differences among treatments. It does not, in any way, prove that all treatments are the same, because the failure to detect treatment differences based on the nonsignificant F test, could be the result of either a very small or no difference among the treatments or due to large experimental error, or both. Thus, whenever the F test is nonsignificant, the researcher should examine the size of the experimental error and the numerical differences among the treatment means. If both values are large, the trial may be repeated and efforts made to reduce the experimental error so that the differences among treatments, if any, can be detected. On the other hand, if both values are small, the differences among treatments are probably too small to be of any economic value and, thus, no additional trials are needed.
For our example, the computed F value of 57.38 is larger than the tabular F value of 3.84 at the 5% level of significance. Hence, the treatment differences are said to be significant. In other words, chances are less than 5 in 100 that all the observed differences among the five treatment means could be due to chance. It should be noted that such a significant F test verifies the existence of some differences among the treatments tested but does not specify the particular pair (or pairs) of treatments that differ significantly. To obtain this information, procedures for comparing treatment means, discussed in Section 4.2.3. are needed.
Step 8. Compute the grand mean and the coefficient of variation (cv) as follows:
Grand mean = ![]() (4.5)
(4.5)
cv = ![]() (4.6)
(4.6)
For our example,
Grand mean = ![]()
cv = ![]()
The cv affects the degree of precision with which the treatments are compared and is a good index of the reliability of the experiment. It is an expression of the overall experimental error as percentage of the overall mean; thus, the higher the cv value, the lower is the reliability of the experiment. The cv varies greatly with the type of experiment, the crop grown, and the characters measured. An experienced researcher, however, can make a reasonably good judgement on the acceptability of a particular cv value for a given type of experiment. Experimental results having a cv value of more than 30 % are to be viewed with caution.
4.2.3. Comparison of treatments
One of the most commonly used test procedures for pair comparisons in forestry research is the least significant difference (LSD) test. Other test procedures, such as Duncanís multiple range test (DMRT), the honestly significant difference (HSD) test and the Student-Newman-Keuls range test, can be found in Gomez and Gomez (1980), Steel and Torrie (1980) and Snedecor and Cochran (1980). The LSD test is described in the following.
The LSD test is the simplest of the procedures for making pair comparisons. The procedure provides for a single LSD value, at a prescribed level of significance, which serves as the boundary between significant and nonsignificant difference between any pair of treatment means. That is, two treatments are declared significantly different at a prescribed level of significance if their difference exceeds the computed LSD value, otherwise they are not considered significantly different.
The LSD test is most appropriate for making planned pair comparisons but, strictly speaking, is not valid for comparing all possible pairs of means, especially when the number of treatments is large. This is so because the number of possible pairs of treatment means increases rapidly as the number of treatments increases. The probability that, due to chance alone, at least one pair will have a difference that exceeds the LSD value increases with the number of treatments being tested. For example, in experiments where no real difference exists among all treatments, it can be shown that the numerical difference between the largest and the smallest treatment means is expected to exceed the LSD value at the 5% level of significance 29% of the time when 5 treatments are involved, 63% of the time when 10 treatments are involved and 83% of the time when 15 treatments are involved. Thus one must avoid use of the LSD test for comparisons of all possible pairs of means. If the LSD test must be used, apply it only when the F test for treatment effect is significant and the number of treatments is not too large, say, less than six.
The procedure for applying the LSD test to compare any two treatments, say the ith and the jth treatments, involves the following steps:
Step 1. Compute the mean difference between the ith and the jth treatment as:
![]() (4.7)
(4.7)
where ![]() are the means of the ith and the jth treatments.
are the means of the ith and the jth treatments.
Step 2. Compute the LSD value at ![]() level of significance as:
level of significance as:
![]() (4.8)
(4.8)
where ![]() is the standard error of the mean difference and
is the standard error of the mean difference and ![]() is the Studentís t value, from Appendix 2, at
is the Studentís t value, from Appendix 2, at ![]() l
evel of significance and with v = Degrees of freedom for error.
l
evel of significance and with v = Degrees of freedom for error.
Step 3. Compare the mean difference computed in Step 1 with the LSD value computed in Step 2 and declare the ith and jth treatments to be significantly different at the ![]() level of significance, if the absolute value of
level of significance, if the absolute value of ![]() is greater than the LSD value.
is greater than the LSD value.
In applying the foregoing procedure, it is important that the appropriate standard error of the mean difference (![]() )for the treatment pair being compared is identified. This task is affected by the experimental design used, the number of replications of the two treatments being compared, and the specific type of means to be compared. In the case of CRD, when the two treatments do not have the same number of replications,
)for the treatment pair being compared is identified. This task is affected by the experimental design used, the number of replications of the two treatments being compared, and the specific type of means to be compared. In the case of CRD, when the two treatments do not have the same number of replications, ![]() is computed as:
is computed as:
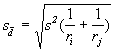 (4.9)
(4.9)
where ri and rj are the number of replications of the ith and the jth treatments and s2 is the error mean square in the analysis of variance.
As an example, use the data from Table 4.1. The researcher wants to compare the five isolates of R. solani, with respect to the mycelial growth on PDA medium. The steps involved in applying the LSD test would be the following.
Step 1. Compute the mean difference between each pair of treatments (isolates) as shown in Table 4.4.
Step 2. Compute the LSD value at a level of significance. Because some treatments have three replications and others have two, three sets of LSD values must be computed.
For comparing two treatments each having three replications, compute the LSD value as follows.
![]()
where the value of s2 = 3.32 is obtained from Table 4.3 and the Studentís t value of 2.31 for 8 degrees of freedom at 5% level is obtained from Appendix 2.
For comparing two treatments each having three replications, compute the LSD value as follows.
![]()
For comparing two treatments one having two replications and the other having three replications, the LSD value is,
![]()
= 3.84 mm
Step 3. Compare difference between each pair of treatments computed in Step 1 to the corresponding LSD values computed in Step 2 and place the appropriate asterisk notation. For example, the mean difference between the first treatment (with three replications) and the second treatment (with three replications) is 2.66 mm. Since the mean difference is less than the corresponding LSD value of 3.44 mm it is declared to be nonsignificant at 5% level of significance. On the other hand, the mean difference between the first treatment (with three replications) and the second treatment (with two replications) is, 4.05 mm. Since the mean difference is higher than the corresponding LSD value of 3.84, it is declared to be significant at the 5% level and is indicated with asterisks. The test results for all pairs of treatments are given in Table 4.4.
Table 4.4. Comparison between mean diameter (mm) of each pair of treatments using the LSD test with unequal replications, for data in Table 4.1.
|
Treatment |
RS 1 |
RS 2 |
RS 3 |
RS 4 |
RS 5 |
|
RS 1 |
0.00 |
2.66 (3.44) |
0.42 (3.84) |
19.33* (3.44) |
4.05* (3.84) |
|
RS 2 |
0.00 |
3.08 (3.84) |
16.67* (3.44) |
1.39 (3.84) |
|
|
RS 3 |
0.00 |
19.75* (3.84) |
4.47* (4.21) |
||
|
RS 4 |
0.00 |
15.28* (3.84) |
|||
|
RS 5 |
0.00 |
* Significant at 5% level
Note: The values in the parenthesis are LSD values
Before leaving this section, one point useful in deciding the number of replications required in an experiment for achieving reasonable level of reliability is mentioned here. As indicated earlier, one thumb rule is to take that many replications which will make the error degrees of freedom around 12. The idea behind this rule is that critical values derived from some of the distributions like Studentís t or F almost stabilize after 12 degrees of freedom thereby providing some extent of stability to the conclusions drawn from such experiments. For instance, if one were to plan a CRD with equal replications for t treatments, one would equate the error df of t(r-1) to 12 and solve for r for known values of t. Similar strategies can be followed for many other designs also that are explained in later sections.
4.3. Randomized complete block design
The randomized complete block design (RCBD) is one of the most widely used experimental designs in forestry research. The design is especially suited for field experiments where the number of treatments is not large and there exists a conspicuous factor based on which homogenous sets of experimental units can be identified. The primary distinguishing feature of the RCBD is the presence of blocks of equal size, each of which contains all the treatments.
4.3.1 Blocking technique
The purpose of blocking is to reduce the experimental error by eliminating the contribution of known sources of variation among the experimental units. This is done by grouping the experimental units into blocks such that variability within each block is minimized and variability among blocks is maximized. Since only the variation within a block becomes part of the experimental error, blocking is most effective when the experimental area has a predictable pattern of variability.
An ideal source of variation to use as the basis for blocking is one that is large and highly predictable. An example is soil heterogeneity, in a fertilizer or provenance trial where yield data is the primary character of interest. In the case of such experiments, after identifying the specific source of variability to be used as the basis for blocking, the size and the shape of blocks must be selected to maximize variability among blocks. The guidelines for this decision are (i) When the gradient is unidirectional (i.e., there is only one gradient), use long and narrow blocks. Furthermore, orient these blocks so that their length is perpendicular to the direction of the gradient. (ii) When the fertility gradient occurs in two directions with one gradient much stronger than the other, ignore the weaker gradient and follow the preceding guideline for the case of the unidirectional gradient. (iii) When the fertility gradient occurs in two directions with both gradients equally strong and perpendicular to each other, use blocks that are as square as possible or choose other designs like latin square design (Gomez and Gomez, 1980).
Whenever blocking is used, the identity of the blocks and the purpose for their use must be consistent throughout the experiment. That is, whenever a source of variation exists that is beyond the control of the researcher, it should be ensured that such variation occurs among blocks rather than within blocks. For example, if certain operations such as application of insecticides or data collection cannot be completed for the whole experiment in one day, the task should be completed for all plots of the same block on the same day. In this way, variation among days (which may be enhanced by weather factors) becomes a part of block variation and is, thus, excluded from the experimental error. If more than one observer is to make measurements in the trial, the same observer should be assigned to make measurements for all plots of the same block. This way, the variation among observers if any, would constitute a part of block variation instead of the experimental error.
4.3.2. Layout
The randomization process for a RCBD is applied separately and independently to each of the blocks. The procedure is illustrated for the case of a field experiment with six treatments A, B, C, D, E, F and three replications.
Step1. Divide the experimental area into r equal blocks, where r is the number of replications, following the blocking technique described in Section 4.3.1. For our example, the experimental area is divided into three blocks as shown in Figure 4.2. Assuming that there is a unidirectional fertility gradient along the length of the experimental field, block shape is made rectangular and perpendicular to the direction of the gradient.
|
Gradient |
|||||
|
Block I |
Block II |
Block III |
|||
Figure 4.2. Division of an experimental area into three blocks, each consisting of six plots, for a randomized complete block design with six treatments and three replications. Blocking is done such that blocks are rectangular and perpendicular to the direction of the unidirectional gradient (indicated by the arrow).
Step2. Subdivide the first block into t experimental plots, where t is the number of treatments. Number the t plots consecutively from 1 to t, and assign t treatments at random to the t plots following any of the randomization schemes for the CRD described in Section 4.2.1. For our example, block I is subdivided into six equisized plots, which are numbered consecutively from top to bottom. (Figure 4.3) and the six treatments are assigned at random to the six plots using the table of random numbers as follows:
|
1 C |
|
2 D |
|
3 F |
|
4 E |
|
5 B |
|
6 A |
|
Block I |
Figure 4.3. Plot numbering and random assignment of six treatments (A, B, C, D, E, and F) to the six plots of Block I.
Step 3. Repeat Step 2 completely for each of the remaining blocks. For our example, the final layout is shown in Figure 4.4.
|
1 |
7 |
13 |
||
|
C |
A |
F |
||
|
2 |
8 |
14 |
||
|
D |
E |
D |
||
|
3 |
9 |
15 |
||
|
F |
F |
C |
||
|
4 |
10 |
16 |
||
|
E |
C |
A |
||
|
5 |
11 |
17 |
||
|
B |
D |
B |
||
|
6 |
12 |
18 |
||
|
A |
B |
E |
||
|
Block I |
Block II |
Block III |
Figure 4.4. A sample layout of a randomized complete block design with six treatments (A, B, C, D, E and F) and three replications.
4.3.3. Analysis of variance
There are three sources of variability in a RCBD : treatment, replication (or block) and experimental error. Note that this is one more than that for a CRD, because of the addition of replication, which c orresponds to the variability among blocks.
To illustrate the steps involved in the analysis of variance for data from a RCBD, data from an experiment is made use of, wherein eight provenances of Gmelina arborea were compared with respect to the girth at breast-height (gbh) of the trees attained since 6 years of planting (Table 4.5).
Table 4.5. Mean gbh (cm) of trees in plots of provenances of Gmelina arborea, 6 years after planting, in a field experiment laid out under RCBD.
|
Treatment (Provenance) |
Replication |
Treatment total |
Treatment mean |
||
|
I |
II |
III |
(Ti) |
|
|
|
1 |
30.85 |
38.01 |
35.10 |
103.96 |
34.65 |
|
2 |
30.24 |
28.43 |
35.93 |
94.60 |
31.53 |
|
3 |
30.94 |
31.64 |
34.95 |
97.53 |
32.51 |
|
4 |
29.89 |
29.12 |
36.75 |
95.76 |
31.92 |
|
5 |
21.52 |
24.07 |
20.76 |
66.35 |
22.12 |
|
6 |
25.38 |
32.14 |
32.19 |
89.71 |
29.90 |
|
7 |
22.89 |
19.66 |
26.92 |
69.47 |
23.16 |
|
8 |
29.44 |
24.95 |
37.99 |
92.38 |
30.79 |
|
Rep. total (Rj) |
221.15 |
228.02 |
260.59 |
||
|
Grand total (G) Grand mean |
709.76 |
29.57 |
|||
Step 1. Group the data by treatments and replications and calculate treatment totals (Ti), replication totals (Rj) and grand total (G), as shown in Table 4.5.
Step 2. Construct the outline of the analysis of variance as follows:
Table 4.6. Schematic representation of ANOVA of RCBD
|
Source of variation |
Degree of freedom (df) |
Sum of squares (SS) |
Mean square
|
Computed F |
|
Replication |
r - 1 |
SSR |
MSR |
|
|
Treatment |
t - 1 |
SST |
MST |
MST/MSE |
|
Error |
(r - 1)(t - 1) |
SSE |
MSE |
|
|
Total |
rt - 1 |
SSTO |
Step 3. Compute the correction factor and the various sums of squares (SS) given in the above table as follows. Let yij represent the observation made from jth block on the ith treatment; i = 1,Ö,t ; j = 1,Ö,r.
C F = ![]() (4.10)
(4.10)
= ![]()
SSTO = ![]() (4.11)
(4.11)
= ![]()
= 678.42
SSR = 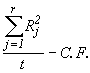 (4.12)
(4.12)
= ![]() 20989.97
20989.97
= 110.98
SST = 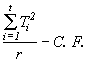 (4.13)
(4.13)
= ![]()
= 426.45
SSE = SSTO - SSR - SST (4.14)
= 678.42 - 110.98 - 426.45 = 140.98
Step 4. Using the values of sums of squares obtained, compute the mean square and the F value for testing the treatment differences as shown in the Table 4.6. The results are shown in Table 4.7.
Table 4.7 ANOVA of gbh data in Table 4.5.
|
Source of |
Degree of |
Sum of |
Mean |
Computed |
Tabular F |
|
variation |
freedom |
Squares |
Square |
F |
5% |
|
Replication |
2 |
110.98 |
55.49 |
|
|
|
Treatment |
7 |
426.45 |
60.92 |
6.05* |
2.76 |
|
Error |
14 |
140.98 |
10.07 |
||
|
Total |
23 |
678.42 |
*Significant at 5% level
Step 5. Obtain the tabular F values from Appendix 3, for f1 = treatment df and f2 = error df. For our example, the tabular F value for f1 = 7 and f2 = 14 degrees of freedom is 2.76 at the 5% level of significance.
Step 6. Compare the computed F value of step 4 with the tabular F values of step 5, and decide on the significance of the differences among treatments. Because the computed F value of 6.05 is greater than the tabular F value at the 5% level of significance, we conclude that the experiment shows evidence the existence of significant differences among the provenances with respect to their growth in terms gbh.
Step 7. Compute the coefficient of variation as:
![]() (4.15)
(4.15)
= ![]()
The relatively low value of cv indicates the reasonable level of precision attained in the field experiment.
4.3.4. Comparison of treatments
The treatment means are compared as illustrated for the case of CRD in Section 4.2.3. using the formulae,
![]() (4.16)
(4.16)
where s-d is the standard error of the difference between treatment means and ![]() is the tabular t value, from Appendix 2, at
is the tabular t value, from Appendix 2, at ![]() level of significance and with v = Degrees of freedom for error. The quantity s-dis computed as:
level of significance and with v = Degrees of freedom for error. The quantity s-dis computed as:
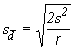 (4.17)
(4.17)
where s2 is the mean square due to error and r is the number of replications.
For illustration, the analysis carried out on data given in Table 4.5 is continued to compare all the possible pairs of treatments through LSD test.
Step 1. Compute the difference between treatment means as shown in Table 4.8.
Table 4.8. Difference between mean gbh (cm) for each pair of treatments of data in Table 4.4.
|
Treatment |
1 |
2 |
3 |
4 |
5 |
6 |
7 |
8 |
|
1 |
0.00 |
3.12 |
2.14 |
2.73 |
12.53* |
4.75 |
11.49* |
3.86 |
|
2 |
0.00 |
0.98 |
0.39 |
9.41* |
1.63 |
8.37* |
0.74 |
|
|
3 |
0.00 |
0.59 |
10.39* |
2.61 |
9.35* |
1.72 |
||
|
4 |
0.00 |
9.8* |
2.02 |
8.76* |
1.13 |
|||
|
5 |
0.00 |
7.78* |
1.04 |
8.67* |
||||
|
6 |
0.00 |
6.74* |
0.89 |
|||||
|
7 |
0.00 |
7.63* |
||||||
|
8 |
0.00 |
* Significant at 5% level
Step 2.Compute the LSD value at a level of significance. Since all the treatments are equally replicated, we need to compute only one LSD value. The LSD value is computed using Equations (4.16) and (4.17).
![]()
Step 3.Compare difference among the treatment means against the computed value of LSD and place the asterisk against significant differences. The results are shown in Table 4.8.
Estimation of missing values
A missing data situation occurs whenever a valid observation is not available for any one of the experimental units. Missing data could occur due to accidental improper application of treatments, erroneous observations, destruction of experimental units due to natural calamities like fire, damage due to wildlife etc. It is extremely important, however, to carefully examine the reasons for missing data. The destruction of the experimental material must not be the result of the treatment e ffect. If a plot has no surviving plants because it has been grazed by stray cattle or vandalized by thieves, each of which is clearly not treatment related, missing data should be appropriately declared. On the other hand, for example, if a control plot (i.e., untreated plot) in an insecticide trial is totally damaged by the insects, the destruction is a logical consequence of that plot being the control plot. Thus, the corresponding plot data should be accepted as valid (i.e., zero yield if all plants in the plot are destroyed, or the actual low yield value if some plants survive) instead of treating it as missing data.
Occurrence of missing data results in two major difficulties; loss of information and non- applicability of the standard analysis of variance. When an experiment has one or more observations missing, the standard computational procedures of the analysis of variance no longer apply except for CRD. One alternative in such cases is the use of the missing data formula technique. In the missing data formula technique, an estimate of a single missing observation is provided through an appropriate formula according to the experimental design used. This estimate is used to replace the missing data and the augmented data set is then subjected, with some slight modifications, to the standard analysis of variance.
It is to be noted that an estimate of the missing data obtained through the missing data formula technique does not supply any additional information, the data once lost is not retrievable through any amount of statistical manipulation. What the procedure attempts to do is to allow the researcher to compute the analysis of variance in the usual manner (i.e., as if the data were complete) without resorting to the more complex procedures needed for incomplete data sets.
A single missing value in a randomized complete block design is estimated as:
![]() (4.18)
(4.18)
where y = Estimate of missing data
t = Number of treatments
r = Number of replications
B0 = Total of observed values of the replication that contains the missing data
T0 = Total of observed values of the treatment that contains the missing data
G0 = Grand total of all observed values
The missing data is replaced by the computed value of y and the usual computational procedure of the analysis of variance is applied to the augmented dataset with some modifications.
The procedure is illustrated with data of Table 4.5, with the value of the sixth treatment (sixth provenance) in replication II assumed to be missing, as shown in Table 4.9. The steps in the computation of the analysis of variance and pair comparisons of treatment means are as follows.
Step 1. Firstly, estimate the missing value, using Equation (4.18) and the values of totals in Table 4.9.
![]() = 26.47
= 26.47
Table 4.9. Data of Table 4.5 with one missing observation
|
Treatment (Provenance) |
Replication |
Treatment total |
||
|
Rep. I |
Rep II |
Rep. III |
(T) |
|
|
1 |
30.85 |
38.01 |
35.1 |
103.96 |
|
2 |
30.24 |
28.43 |
35.93 |
94.6 |
|
3 |
30.94 |
31.64 |
34.95 |
97.53 |
|
4 |
29.89 |
29.12 |
36.75 |
95.76 |
|
5 |
21.52 |
24.07 |
20.76 |
66.35 |
|
6 |
25.38 |
M |
32.19 |
(57.57=T0) |
|
7 |
22.89 |
19.66 |
26.92 |
69.47 |
|
8 |
29.44 |
24.95 |
37.99 |
92.38 |
|
Rep. total (R) |
221.15 |
(195.88=B0) |
260.59 |
|
|
Grand total (G) |
(677.62=G0) |
|||
M = Missing data
Step 2. Replace the missing data of Table 4.9. by its estimated value computed in step 1, as shown in Table 4.10 and carry out the analysis of variance of the augmented data set based on the standard procedure of Section 4.3.3.
Table 4.10. Data in Table 4.7 with the missing data replaced by the value estimated from the missing data formula technique.
|
Treatment (Provenance) |
Replication |
Treatment total |
||
|
Rep. I |
Rep II |
Rep. III |
(T) |
|
|
1 |
30.85 |
38.01 |
35.1 |
103.96 |
|
2 |
30.24 |
28.43 |
35.93 |
94.6 |
|
3 |
30.94 |
31.64 |
34.95 |
97.53 |
|
4 |
29.89 |
29.12 |
36.75 |
95.76 |
|
5 |
21.52 |
24.07 |
20.76 |
66.35 |
|
6 |
25.38 |
26.47a |
32.19 |
84.04 |
|
7 |
22.89 |
19.66 |
26.92 |
69.47 |
|
8 |
29.44 |
24.95 |
37.99 |
92.38 |
|
Rep. total (R) |
221.15 |
222.35 |
260.59 |
|
|
Grand total (G) |
704.09 |
|||
a Estimate of the missing data obtained from missing data formula technique
Step 3. Make the following modifications to the analysis of variance obtained in Step 2; Subtract 1 from both the total and error df. For our example, the total df of 23 becomes 22 and the error df of 14 becomes 13. Compute the correction factor for bias (B) as,
B = 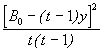 (4.19)
(4.19)
= ![]()
= 2.00
and subtract the computed B value of 2.00 from the treatment sum of squares and the total sum of squares. For our example, the SSTO and the SST, computed in Step 2 from the augmented data of Table 4.10, are 680.12 and 432.09, respectively. Subtracting the B value of 2.00 from these SS values, we obtain the adjusted SST and the adjusted SSTO as:
Adjusted SST = 432.09 - 2.00
= 430.09
Adjusted SSTO = 680.12 - 2.00
= 678.12
The resulting ANOVA is shown in Table 4.11.
Table 4.11. Analysis of variance of data in Table 4.7 with one missing value estimated by the missing data formula technique.
|
Source of |
Degree of |
Sum of |
Mean |
Computed |
Tabular F |
|
variation |
freedom |
squares |
square |
F |
5 % |
|
Replication |
2 |
125.80 |
62.90 |
6.69 |
|
|
Treatment |
7 |
430.09 |
61.44 |
6.53* |
2.83 |
|
Error |
13 |
122.23 |
9.40 |
||
|
Total |
22 |
678.12 |
* Significant at 5% level of significance
Step 4. For pairwise comparisons of treatment means where one of the treatments has missing data, compute the standard error of the mean difference ![]() as:
as:
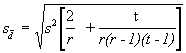 (4.20)
(4.20)
where s2 is the error mean square from the analysis of variance of Step 3, r is the number of replications, and t is the number of treatments.
For example, to compare the mean of the sixth treatment (the treatment with missing data) with any one of the other treatments, s-d is computed as:
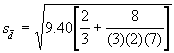 = 2.84
= 2.84
This computed s-d is appropriate for use in the computation of the LSD values. For illustration, the computation of the LSD values is shown below. Using tv as the tabular t value for 13 df at 5% level of significance, obtained from Appendix 3, the LSD values for comparing the sixth treatment mean with any other treatment mean is computed as:
LSDa
=tv;a![]() (4.21)
(4.21)
LSD.05 = (2.16)(2.84) = 6.13