Growth and/or inactivation of pathogenic and spoilage microorganisms are very important factors determining safety and shelf-life of seafood. Clearly, assessment and management of safety and quality is facilitated when microbial growth and inactivation can be quantitatively related to characteristics of products and processes like temperature, atmosphere, pH and NaCl %. Predictive microbiology is the area of food microbiology where such relations between controlling factors in foods and responses of pathogenic and spoilage microorganisms are quantified and modelled by mathematical equations. Predictive microbiology has numerous practical applications and is an active area of research.
Large amounts of experimental data are required to predict the effect of controlling factors on growth, probability of growth, survival or inactivation of microorganisms (Table 14.1). Such data have often been generated using liquid laboratory media as levels of controlling factors (pH, NaCl %, etc.) are easy to adjust. In addition, automated methods for measuring microbial growth such as absorbance or conductance measurements can be used to facilitate the generation of data in liquid media. However, to accurately predict microbial growth in seafoods, liquid media cannot be used uncritically. With apparently similar levels of controlling factors, growth rates in seafood and standard liquid media like Brain Heart Infusion or Tryptone Soya Broth may differ by a factor of two. Therefore generation of data in product storage trials can be required for development of accurate predictive models (Dalgaard et al., 2002). Knowledge about controlling factors is a prerequisite for development of accurate predictive models. In fact, major controlling factors and even the microorganisms responsible for seafood spoilage have in some cases remained unknown until mathematical modelling studies was initiated.
Table 14.1 Summary of general methodology for development of predictive growth models.
|
Data generation |
Primary modelling |
Secondary modelling |
Product validation |
|
Generate growth curves for combinations of controlling factors (temp., atmosphere, pH, etc.). Often liquid laboratory media are used. |
Estimate kinetic parameters, primarily lag time and maximum specific growth rate, by fitting of growth curves with appropriate model |
Model the effect of controlling factors on lag phase and growth rates |
Evaluate the performance of a model by comparison of predictions and kinetic parameter values determined in product studies |
To estimate lag times, maximum specific rates of growth (µmax) or rates of inactivation, simple primary models are available and include:
(i) the exponential model, with or without a lag phase (Lodge and Hinshelwood 1943)
(ii) the three parameter Logistic model (Eqn. 14.1; solid line in Figure 14.1) or
(iii) four parameter versions of the Logistic model (Eqn. 14.2).
Numerous more flexible and complex primary models have been suggested but in most cases they have no advantage over the simpler primary growth models (Dalgaard, 2002). Nevertheless, the practical usefulness of at least one of the more complicated primary growth models has been increased by including it in the MicroFit software which is available free of charge (www.ifr.bbsrc.ac.uk/MicroFit/).
|
|
14.1 |
|
|
14.2 |
In eqn. 14.1 and 14.2 Nt is the cell concentrations (cfu g-1) at the time t, Nmax and Nmin, respectively, are the maximum and minimum cell concentrations (cfu g-1), µmax the maximum specific growth rate (h-1) and ti the time when Nt = Nmax/2 i.e. the inflection point.
Microbial interactions can be an important factor controlling growth of pathogenic microorganisms in seafood. E.g. for sliced and vacuum packed cold-smoked salmon, growth of Listeria monocytogenes has been found to cease when lactic acid bacteria reach their maximum cell concentration (Figure 14.1). This so-called Jameson effect can be modelled by a simple expansion of the differential form of the Logistic model (Eqn. 14.3, Jørgensen, 2000; Ross et al., 2000a; Dalgaard, 2002).
|
Figure 14.1 |
|
|
|
14.3 |
In eqn. 14.3 Lm and LAB signifies lactic acid
bacteria and L. monocytogenes, respectively. dLm/dt is the
absolute growth rates, Lmt and LABt cell
concentrations (cfu g-1) at the time t, Lmmax
and LABmax the maximum cell contrations (cfu
g-1) and 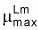 the
maximum specific growth rate (h-1).
the
maximum specific growth rate (h-1).
Polynomial equations have been used extensively as secondary models for estimating the combined effect of several controlling factors on values of lag time and maximum specific growth rates (McClure et al. 1994). The polynomial models include a relatively large number of parameters and this makes it difficult to compare values from different studies particularly as the parameters have no biological interpretation. In contrast, square root type models like eqn. 14.4 include parameters with some biological meaning. The parameters Tmin, aw min, pHmin and %CO2 max correspond to theoretical growth limits for temperature, water activity, pH and CO2. Rather than corresponding to the lowest temperature, water activity, pH or the highest CO2 level where growth is actually observed Tmin, aw min, pHmin or %CO2 max are determined mathematically as the extrapolated values where the growth rate as a function of these controlling factors theoretically becomes zero. Thus, Tmin -values of -5°C to -10°C are common for psychrotolerant Gram-negative bacteria although these microorganisms are typically inactivated at -5°C to -10°C. Nevertheless, Tmin, aw min, pHmin or %CO2 max each seems little influenced by other controlling factors. Therefore, when reliable estimates of these parameters are available, only few data are required to develop new models for specific pathogen/product combinations. As an example a model for growth of L. monocytogenes in a specific seafood may be developed from values of existing parameters and storage trials required to estimate the value of 'b' in eqn. 13.4 (Ross et al., 2000a). This approach has not yet been extensively used and deserves further study. Table 14.2 shows values of Tmin, aw min, pHmin or %CO2 max for selected pathogenic and spoilage bacteria of importance in seafood.
|
|
14.4 |
Irrespective of the approach and the type of equations applied, development of a predictive model must always include product validation studies to evaluate the performance of the model. Graphs showing observed and predicted values of lag times, maximum specific growth rates or times for e.g. a 1000-fold increase in cell concentrations are useful to evaluate the performance of predictive growth models. In addition, the bias factor (Eqn. 14.5) and accuracy factor (Eqn. 14.6) are most important indices of performance for predictive models (Ross, 1996).
Table 14.2 Values of Tmin, aw min, pHmin and %CO2 max for selected pathogenic and spoilage bacteria of importance in seafood. Data from Ross and McMeekin 1991, Miles et al. 1997, Presser et al. 1997, Ross et al. 2000a, Dalgaard 2002.
|
|
Tmin |
aw min |
pHmin |
% CO2 max |
|
Escherichia coli |
+4.0 |
0.93 |
3.9 |
- |
|
Listeria monocytogenes |
0 |
0.92 |
4.2 |
- |
|
Staphylococcus aureus |
+7.4 |
0.87 |
- |
- |
|
Vibrio parahaemolyticus |
+5.4 |
0.92 |
- |
- |
|
Brochothrix thermosphacta |
-10.9 |
- |
- |
187* |
|
Lactobacillus curvatus |
-3.3 |
0.93 |
4.2 |
- |
|
Photobacterium phosphoreum |
-9.0 |
0.95 |
4.3 |
376 |
|
Shewanella putrefaciens |
-8.0 to -9.9 |
0.95 |
- |
150-156 |
* Values of CO2 above 100% correspond to partial pressures above atmospheric pressure.
|
|
14.5 |
|
|
14.6 |
The bias factor indicates systematic over- or under prediction and a value of 1.0 shows predicted and observed values to be equal on average. A bias factor value between 0.75 and 1.25 has been suggested as a criterion for successful validation of microbial models to predict shelf-life of seafood. For pathogenic microorganisms limits for bias factors has been suggested to be slightly closer to 1.0 (Dalgaard, 2000; Ross et al., 2000a).
|
Bias factor Index of performance to compare predicted growth and values observed in product studies. Successful validation of a predictive model requires a bias factor value between 0.75 and 1.25 for a specific seafood |
Predictive models developed in liquid laboratory media may not include all major factors actually limiting microbial growth in seafood. Predictions from such incomplete models can be strongly biased and if used uncritically predictions can be misleading. As an example, models including the effect of temperature, NaCl/aw, pH and lactate were unable to accurately predict growth of L. monocytogenes in naturally contaminated cold-smoked salmon. In fact, the bias factor was above 5 and it was pointed out that the effect of microbial interactions and smoke components was missing in the existing models (Dalgaard and Jørgensen, 1998; Ross et al., 2000a). Recent studies confirmed that predictions could be substantially improved when the effect of interaction between L. monocytogenes and lactic acid bacteria (Eqn. 14.3) was added to a model already including the effect of temperature, NaCl/aw, pH and lactate (Ross et al., 2000b; Dalgaard, 2002).
Experimental data continuously indicate that the major factors controlling microbial growth in seafood are not all identified. Clearly, predictive models may be incomplete and should never be used uncritically. Users of a model must verify that a bias factor between 0.75 and 1.25 has been obtained in product validation studies before predictions are applied for assessment or management of seafood safety. Most important, the validation studies need to be carried out with seafood having microbial ecology similar to the product of interest. Furthermore, if a bias factor is e.g. 1.4 and a single controlling factor like smoke components in cold-smoked salmon is lacking in the model, then predictions can be corrected by the value of bias factor. In this way, a corrected model can be used (with caution) to predict the effect of the factors actually included in the model.
Successfully validated predictive models have numerous applications in assessment and management of seafood safety and quality, particularly when models are included in application softwares. Application software does not improve the models but they allow users, including people without interest in the mathematics of microbial kinetics, to obtain prediction rapidly and conveniently.
Seafood is never distributed at a constant temperature and in practice fresh and lightly preserved products that are supposed to be chilled can be exposed to between ~ 0°C and ~ 15°C. In tropical regions the temperature of fish raw material may be in the range of 25-30°C. Thus, the rate of chilling is a very critical parameter for both shelf-life and safety of products. The Seafood Spoilage Predictor (SSP) software has been developed specifically to predict the effect of constant and fluctuating temperature conditions on shelf-life of products from temperate and tropical waters as well as on growth of the spoilage bacteria Photobacterium phosphoreum and Shewanella puftefaciens. SSP is available free of charge at www.dfu.min.dk/micro/ssp/ (Dalgaard et al., 2002). The Food Spoilage Predictor (FSP) software (www.geminidataloggers.com) and several predictive models are available (Koutsoumanis 2001, Rasmussen et al. 2002) for prediction of growth of psychrotolerant pseudomonads in fresh aerobically stored fish.
Pathogen Modeling Programme (PMP, www.arserrc.gov/mfs/PATHOGEN.HTM) includes 13 models for growth and survival and 9 models for inactivation of pathogenic bacteria. Food MicroModel (Anon., 1997) includes 23 growth and survival models and 7 models for heat inactivation of primarily pathogenic microorganisms. These software packages do not yet allow models to be used for time-temperature integration as described above. Models in PMP and Food MicroModel include the effect of wide ranges of several controlling factors. Thus, the models may be used to determine how one controlling factors e.g. NaCl % can be substituted by other factors such as a reduction of temperature or a change in packaging from aerobic to vacuum or modified atmosphere. Models in PMP and Food MicroModel may also be used to establish limits for critical control point as part of HACCP plans. Unfortunately, successful validation of the growth, survival and inactivation models in PMP and Food MicroModel remain to be documented for most types of seafood.
For L. monocytogenes in cold-smoked salmon and Vibrio parahaemolyticus and V. vulnificus in raw oysters predictive models have recently been included in exposure assessment models. By using the predictive models together with Monte Carlo simulation software the effect of initial product contamination, product temperature and product characteristics on levels of the pathogens can be predicted at the time product are consumed. Used in this way predictive models become a key component in quantitative risk assessments (Ross et al., 2000b; FAO/WHO, 2002).
In the future, the use of predictive models in the assessment and management of seafood safety and quality will most likely increase substantially. New software to predict safety and shelf-life is likely to appear and predictive models may be combined with seafood traceability systems.
References
Anonymous 1997. Food MicroModel - User Manual v. 2.5. Food Micromodel Ltd., Surrey, UK.
Dalgaard, P. 2000. Fresh and lightly preserved seafood. In: Man,C.M.D. and A.A. Jones (eds) Shelf-Life Evaluation of Foods. Aspen Publishers Inc., London, UK. pp. 110-139.
Dalgaard, P. 2002. Modelling and prediction the shelf-life of seafood. In: Bremner, H.A. (ed.) Safety and quality issues in fish processing. Woodhead Publishing Ltd. pp. 191-219.
Dalgaard, P. and L.V. Jørgensen 1998. Predicted and observed growth of Listeria monocytogenes in seafood challenge tests and in naturally contaminated cold smoked salmon. International Journal of Food Microbiology 40, 105-115.
Dalgaard, P., P. Buch and S. Silberberg 2002. Seafood Spoilage Predictor - development and distribution of a product specific application software. International Journal of Food Microbiology 73, 227-233.
FAO/WHO (Food and Agriculture Organization/World Health Organization) 2002. Joint FAO/WHO Activity on Risk Assessment of Microbiological Hazards in Foods. Hazard identification, exposure assessment and hazard characterizatrion of Vibrio spp. in seafood. Preliminary document.
Jørgensen, L. V. 2000. Spoilage and safety of cold-smoked salmon. Ph.D. thesis. Danish Institute for Fisheries Research, Lyngby, and the Royal Veterinary and Agricultural University, Frederiksberg, Denmark.
Koutsoumanis, K. 2001. Predictive modeling of the shelf life of fish under nonisothermal conditions. Applied and Environmental Microbiology 67, 1821-1829.
Lodge, R.M. and C.N. Hinshelwood 1943. Physiological aspects of bacterial growth. Part IX. The lag phase of Bact. Lactis Aerogenes. Journal of the Chemical Society 288, 213-219.
McClure, P.J., C.D. Blackburn, M.B. Cole, P.S. Curtis, J.E. Jones, J.D. Legan, I.D. Ogden and M.W. Peck 1994. Modelling the growth, survival and death of microorganisms in foods: the UK Food Micromodel approach. International Journal of Food Microbiology 23, 265-275.
Miles, D.W., T. Ross, J. Olley and T.A. McMeekin 1997. Development and evaluation of a predictive model for the effect of temperature and water activity on the growth rate of Vibrio parahaemolyticus. International Journal of Food Microbiology 38, 133-142.
Presser, K.A., D.A. Ratkowsky and T. Ross 1997. Modelling the growth rate of Escherichia coli as a function of pH and lactic acid concentration. Applied and Environmental Microbiology 63, 2355-2360.
Rasmussen, S.K.J., T. Ross, J. Olley and T.A. McMeekin 2002. A process risk model for the shelf-life of Atlantic salmon fillets. International Journal of Food Microbiology 73, 47-60.
Ross, T. 1996. Indices for performance evaluation of predictive models in food microbiology. Journal of Applied Bacteriology 81, 501-508.
Ross, T.A. and T.A. McMeekin 1991. Predictive microbiology: Application of a square root model. Food Australia 43, 202-207.
Ross, T., P. Dalgaard and S. Tienungoon 2000a. Predictive modelling of the growth and survival of Listeria in fishery products. International Journal of Food Microbiology 62, 231-245.
Ross, T., E. Todd and M. Smith 2000b. Exposure assessment of L. monocytogenes in ready-to-eat foods. World Health Organization, Geneva, and Food and Agriculture Organization of the United Nations, Rome, Italy.