by
TERENCE M. THOMAS and CLIFFORD L. FISHBACK
Institute of Marine Sciences
University of Miami
Miami, Florida 33149, U.S.A.
Abstract
A computer program is presented to summarize ecological data and to estimate the sample size necessary for establishing an index of abundance from replicate samples. The dimensions of these programs are 10 replicates and 999 individuals per sample. The writing of summary tables is done with an autocoder program compatible with a 1401 computer. Calculations of the sample variance, coefficient of variation and number of replicates necessary to estimate relative abundance are made with a Fortran II program which is compatible with most computer systems.
PROGRAMMATION DE CALCULATEURS POUR LE TRAITEMENT DES DONNEES SUR L'ABONDANCE DES CREVETTES ET ESPECES AFFINES
Résumé
Les auteurs présentent un programme pour calculateur électronique visant à récapituler les données écologiques et à estimer la taille de l'échantillon nécessaire pour établir un indice d'abondance à partir d'échantillonnages réitérés. Dimensions du programme: 10 réitérations et 999 individus par échantillon. Les tableaux récapitulatifs sont établis à l'aide d'un programme autocodeur compatible avec un ordinateur 1401. Le calcul de la variance de l'échantillon, du coefficient de variation et du nombre de réitérations nécessaires pour l'estimation de l'abondance relative est fait au moyen d'un programme Fortran II, compatible avec la plupart des systèmes de calculateurs.
PROGRAMA CON CALCULADORAS PARA TRATAR LOS DATOS SOBRE LA ABUNDANCIA DE CAMARONES Y ANIMALES ASOCIADOS
Extracto
Se expone un programa con calculadoras para resumir datos ecológicos y determinar el tamaño de la muestra necesario para poder establecer un índice de la abundancia mediante muestras reiteradas. Las dimensiones de estos programas son de 10 reiteraciones y 999 ejemplares por muestra. La elaboración de los cuadros resumidos se efectúa con un programa autocodificador compatible con una calculadora 1401. Los cálculos de la variancia de la muestra, coeficiente de variación y número de reiteraciones precisas para estimar la abundancia relativa se realizan con un programa Fortran II que es compatible con casi todos los sistemas de calculadoras.
In September 1963, the Division of Fishery Sciences of the Institute of Marine Sciences, University of Miami, began a quantitative ecological study of the estuarine regions of Everglades National Park. Particular emphasis was placed on assessing the effects of fresh water supply upon the biota, of which the pink shrimp, Penaeus duorarum Burkenroad is a dominant member. This study was supported by U.S. Public Health Service, Division of Water Supply and Pollution Control.
The principal aims of the investigation were to relate seasonal variations at the species and population levels to changes in salinity, temperature and substrate type. Other parameters were also considered and provisions were made in establishing the computer program to include eight additional habitat variables in subsequent computer analyses.
Tagging of juvenile pink shrimp P. duorarum in the Buttonwood Canal and Shark River areas (Tabb, Dubrow and Jones, 1962; Costello and Allen, 1966) has demonstrated a link between this population and the shrimp caught in the Tortugas fishery (Fig. 1). Water in the study area undergoes seasonal salinity fluctuations between 1 and 70‰, providing an opportunity to study changes in abundance and size composition of the shrimp and other populations in response to salinity changes.
The sampling method and the statistical treatment of the data are described in detail by Tabb and Thomas (MS) and are summarized briefly here. The first year of the 3-yr study was devoted to the development of a quantitative method of sampling. Potential sources of variation such as towing speed, towing direction and substrate type were evaluated. Replicate samples were taken from a permanent station of 100 yd2 (91.4m2), near Murray Key (Fig. 1). A fixed station was chosen rather than operating in a larger area on a random sampling basis because the study of effects of environmental fluctuations on animal abundance was the major consideration (Greig-Smith, 1964). The substrate of the sampling area consisted of calcium carbonate mud covered by a dense growth of turtle grass, Thallassia testudinum. The sampling gear was a 2-m otter trawl (modified shrimp “try net”) from Standard Marine Supply, Key West, Florida. A ½-in stretched mesh liner was inserted in the codend of the net.
Samples were collected once every two weeks for a period of one year. Ten drags per trip were chosen because that was the greatest number of samples that could be sorted conveniently during the two-week period between trips.
The data were analyzed to determine how many samples were required to detect changes in the population mean abundance. Estimates of the required number of replicates (N) were made using 10 months' data. The maximum allowable error was ±50 percent of the population mean. This error was tolerated to allow a large number of species to serve as possible environmental indicators.
The number of replicates (N) needed to obtain estimates which are within ±50 percent of the mean were obtained from the formula
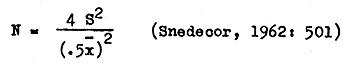
The long series of replicated observations collected during this study required mechanical data handling. The system described below is compatible with an IBM 1401 computer.
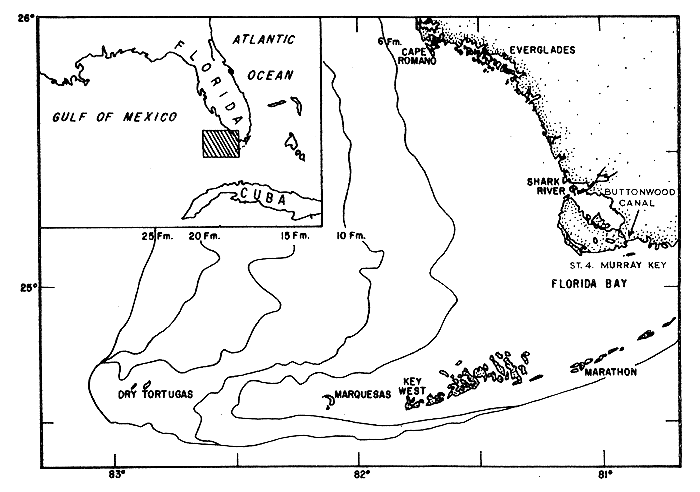
Fig. 1 The location of the sampling site (Station 4) in relation to the estuarine areas of Everglades National Park and the Tortugas fishing grounds.
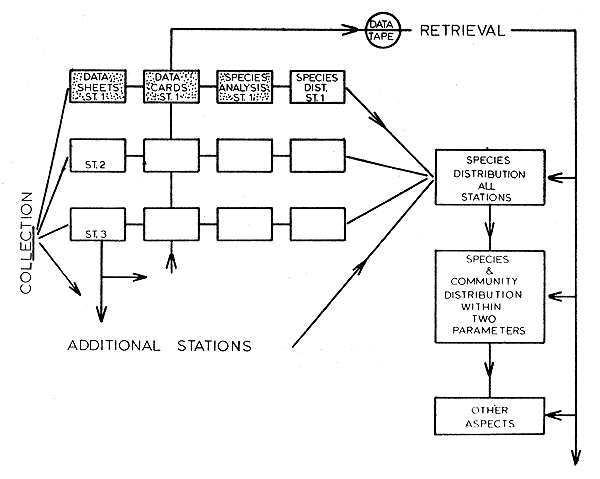
Fig. 2 Generalized flow chart of the complete data processing system. The stippled area represents that portion explained in this paper.
A generalized flow chart of the data storage and retrieval system developed for this study appears in Fig. 2. The stipled portion of this figure represents the part of the system used to analyze the shrimp data presented in this paper. However, whether utilizing the whole or only part of this system, the methods of data acquisition, recording, and punched card formats remain the same. Standardization of these initial stages does not result in the loss of any flexibility, and the system remains open-ended at all times. No compensatory measures are needed to offset seasonal changes in species composition at the station level, and the number of stations may be increased or decreased at any time.
Careful consideration of the purposes of the experiment will make it easier to organize the data on the card. In the same way, field data sheets can be designed which minimize the work in transferring data. Where the initial goals of the experiment are less clear, more complicated data sorting procedures may be required.
The field data sheet designed for this program is shown in Fig. 3. The upper portion of this form, under the sub-heading “Samples”, provides enough space to record the data of up to ten replicate samples. Space is provided to include the measurements of 50 individual animals from each of the 10 samples, while the total number of individuals per sample is recorded in the appropriate space under the sub-heading “Punch”. In this section, a “minus one” was included if samples were not taken or were lost. This provides the computer with the necessary information to adjust for a change of sample size when making other calculations. The sample weight and the average size, minimum size and maximum size of individuals can also be filled in by the person sorting the sample. The rest of the information on this line is explained in detail in the appendix section on data card format.
On completion, these data sheets can be given directly to a key punch operator and data cards cut from them. One data sheet per species per trip is required.
The input for the program consists of punched cards. Three specific card types are used to make up the library file. These are:
Name card (scientific name, code number);
Data card (how many, what, where, when, code number); and
Parameter card (environmental conditions, where, when, code number).
Although only one species is being considered in this paper, complete cross referencing is possible for any number of species and stations by the use of appropriate code numbers on each card. The data card is the control (e.g., if no data exist for a certain species on a certain trip, then there is no utilization of the name or parameter cards). Such a coded cross reference system allows for a greater condensation of information, either in core storage or on tape. This, in turn, provides an economy of time and effort by avoiding unnecessary duplication. Specific details of the card format for each card type are shown in the Appendix Tables I, II and III.
5.1 Sorting routine
All input data must be in some logical order acceptable to the program being used. With this program the sequence is as follows. The three kinds of cards listed earlier are sorted by ascending species code number, followed by an ascending order of each trip, each year and each station separately. In order to make this separation of the data when using an IBM 082 card sorting machine, the order of sorting by card columns must be 70, 69, 68, 67, 75, 74, 73, 72, 76, 78, 80.

Fig. 3 The field data sheet utilized in this study. Sizes of individual animals are recorded for each sample under the heading Samples. The two circled numbers are the maximum and minimum lengths. Information recorded under the heading Punch is the required data card format and includes the total number caught per sample.
The program required to carry out the analysis represented by the stippled area in Fig. 2 is a three-phase operation. A summary of each phase is given in Table I.
A program to operate phase 1 has not been given in this paper since this step can be accomplished by using a standard utility program (i.e., cards to tape). The source programs for phases 2 and 3 are appended (Appendix Tables IV and V). The final printed output from phase 3 of this program is given in Table II. The layout of this table is identical to the actual computer printout.
TABLE I
Summary of the input, operation and output steps required to produce the printed output of Table II
| Phase | Input | Operation | Output | |
| 1 | Cards - | Data | Cards to tape | Tape 1 Data |
| Parameter | Utility program not listed | Tape 2 Parameters | ||
| Name | Tape 3 Names | |||
| 2 | Tape 1 of output phase 1 | Calculations | Tape 4 | |
| Fortram program (Appendix Table IV) | ||||
| 3 | Tape 4 | Construction of arrays Autocoder program (Appendix Table V) | Printed (Listing Table II) | |
| Tape 2 | ||||
| Tape 3 | ||||
The program described in this paper provides a complete and condensed presentation by species by station of data collected in a large scale ecological investigation. Ten environmental parameters can be included.
This program is open-ended, permitting changes in the number of species or stations. It can be applied to any set of replicate samples providing these do not exceed ten. If the number of individuals per sample exceeds the maximum field size of 999, a suitable scale can be applied to the data. However, other card field sizes (i.e. parameters, codes, etc.) must be adhered to (see Appendix Tables I, II and III).
The calculation of the number of replicates (N), the sample variance, the standard deviation and the coefficient of variation provide the basic statistics. The layout of the printed output in Table II is compatible with the input requirements of several multi-variate statistical programs (Clyde, Cramer and Sherin, 1966) if a more sophisticated analysis of the data is required.
The use of a computer program like the one described here speeds up the analysis of data in suitable cases. But considerable work is required to reorganize data into the necessary formats unless proper field data sheets are designed.
TABLE II
Computer printout of the first ten months data, consisting of 20 trips with 10 replicate samples per trip. * Data columns left to right: Date, salinity in parts per thousand, Temperature in °C, time of day, wind direction and velocity, vegetation weight, tunicate weight, sponge weight, precipitation between trips, evaporation between trips, species weight in grams, mean length in millimeters, minimum length in millimeters, maximum length in millimeters, total number of shrimp, mean number of shrimp, sample variance, standard deviation, coefficient of variation, detectable error ± 50% mean squared × two, estimated number of replicates (N), station number.
| Yearly Analysis | Jun. 25, 1965 | Page 21 | |||||||||||||||||||||
| Decapoda (Penaeidae) Penaeus duorarum | 0210C 4 | ||||||||||||||||||||||
| Parameters | Variables | ||||||||||||||||||||||
| Date | Sal. | Temp | Time | Wind | VG.W.T. | T.WT. | S.WT. | Prec. | Evap. | WT. | Size | Min | Max | Sum | Mean | Var. | Dev. | C.V. | D.F. | N | S | ||
| 12 | Feb 64 | 33.9 | 16.5 | 1430 | NE | 5.4 | 12,984 | 1,566 | 0.64 | 2.43 | 232.1 | 12.3 | 9 | 15 | 4 | 0.4 | 0.5 | 0.7 | 1.7 | 0.1 | 24.4 | 4 | |
| 10 | Mar 64 | 35.4 | 26.0 | 1255 | S | 7.8 | 23,789 | 0.02 | 2.51 | 459.3 | 13.4 | 8 | 20 | 85 | 8.5 | 30.1 | 5.5 | 0.6 | 36.1 | 3.3 | 4 | ||
| 24 | Mar 64 | 35.6 | 22.0 | 1235 | SE | 7.7 | 21,973 | 1,475 | 1.49 | 3.62 | 212.8 | 13.5 | 11 | 17 | 30 | 3.0 | 3.1 | 1.8 | 0.6 | 4.5 | 2.8 | 4 | |
| 7 | Apr 64 | 39.8 | 24.4 | 1355 | SE | 8.0 | 23,744 | 846 | 0000 | 2.85 | 215.4 | 14.6 | 9 | 19 | 16 | 1.6 | 3.4 | 1.8 | 1.1 | 1.3 | 10.6 | 4 | |
| 22 | Apr 64 | 39.8 | 25.4 | 1320 | E | 5.9 | 45,400 | 910 | 0.06 | 4.09 | 266.6 | 15.3 | 9 | 20 | 9 | 0.9 | 0.5 | 0.7 | 0.8 | 0.4 | 5.4 | 4 | |
| 5 | May 64 | 41.0 | 25.4 | 0955 | NE | 5.3 | 29,918 | 1,038 | 1.64 | 2.42 | 287.8 | 8.7 | 3 | 17 | 13 | 1.3 | 3.8 | 1.9 | 1.5 | 0.8 | 17.9 | 4 | |
| 19 | May 64 | 42.0 | 24.6 | 1007 | NE | 5.0 | 54,934 | 756 | 0.51 | 3.54 | 592.2 | 7.9 | 4 | 12 | 22 | 2.2 | 2.4 | 1.5 | 0.7 | 2.4 | 4.0 | 4 | |
| 3 | Jun 64 | 40.2 | 26.6 | 0938 | SW | 2.7 | 42,630 | 634 | 12.50 | 4.26 | 2.44 | 548.5 | 8.6 | 3 | 15 | 104 | 10.4 | 8.0 | 2.8 | 0.3 | 54.1 | 0.6 | 4 |
| 17 | Jun 64 | 38.2 | 28.1 | 0932 | E | 3.1 | 14,934 | 518 | 14.50 | 8.04 | 2.50 | 238.5 | 9.6 | 4 | 36 | 38 | 3.8 | 5.3 | 2.3 | 0.6 | 7.2 | 2.9 | 4 |
| 2 | Jul 64 | 39.3 | 28.2 | 0845 | E | 3.3 | 6,341 | 684 | 1.57 | 3.38 | 686.9 | 9.5 | 5 | 16 | 31 | 3.1 | 4.8 | 2.2 | 0.7 | 4.8 | 4.0 | 4 | |
| 16 | Jul 64 | 39.6 | 27.2 | 0839 | SE | 4.2 | 3,586 | 1,185 | 3.10 | 0.99 | 3.84 | 251.9 | 12.5 | 8 | 18 | 11 | 1.1 | 1.2 | 1.1 | 1.0 | 0.6 | 8.0 | 4 |
| 31 | Jul 64 | 42.4 | 29.0 | 0730 | NE | 7.9 | 5,584 | 965 | 37.00 | 1.78 | 3.81 | 184.7 | 11.1 | 8 | 16 | 12 | 1.2 | 1.1 | 1.0 | 0.9 | 0.7 | 5.9 | 4 |
| 8 | Aug 64 | 41.6 | 29.5 | 1630 | SE | 4.3 | 4,630 | 941 | 37.90 | 1.17 | 1.81 | 193.0 | 11.3 | 7 | 18 | 22 | 2.2 | 15.3 | 3.9 | 1.8 | 2.4 | 25.3 | 4 |
| 24 | Aug 64 | 45.0 | 30.8 | 1412 | E | 3.1 | 3,314 | 800 | 96.80 | 1.07 | 4.24 | 168.6 | 10.1 | 4 | 20 | 28 | 2.8 | 3.7 | 1.9 | 0.7 | 3.9 | 3.8 | 4 |
| 9 | Sep 64 | 41.6 | 29.9 | 1615 | W | 5.1 | 2,315 | 874 | 36.40 | 2.02 | 3.38 | 1727.3 | 11.7 | 7 | 21 | 258 | 25.8 | 50.2 | 7.1 | 0.2 | 332.8 | 0.6 | 4 |
| 29 | Sep 64 | 40.3 | 28.5 | 0735 | SE | 4.0 | 2,133 | 436 | 31.70 | 3.99 | 2.21 | 349.2 | 9.0 | 7 | 11 | 3 | 0.3 | 0.2 | 0.5 | 1.6 | 0.0 | 20.7 | 4 |
| 20 | Oct 64 | 37.2 | 22.5 | 1330 | N | 1.7 | 7,127 | 966 | 24.50 | 2.21 | 2.63 | 1612.2 | 12.6 | 7 | 23 | 543 | 54.3 | 475.1 | 21.8 | 0.4 | 1474.2 | 1.3 | 4 |
| 17 | Nov 64 | 33.4 | 25.3 | 1330 | NE | 2.1 | 6,004 | 770 | 3.27 | 3.95 | 289.5 | 13.1 | 10 | 17 | 61 | 8.7 | 55.9 | 7.5 | 0.9 | 38.0 | 5.9 | 4 | |
| 15 | Dec 64 | 33.7 | 21.2 | 1130 | NE | 3.8 | 8,896 | 595 | 0.32 | 3.06 | 3461.0 | 14.9 | 6 | 28 | 858 | 122.6 | 481.0 | 21.9 | 0.2 | 7511.9 | 0.3 | 4 | |
| 14 | Jan 65 | 33.4 | 23.0 | 1230 | SE | 1.8 | 7,593 | 769 | 0.31 | 4.39 | 458.7 | 15.1 | 10 | 20 | 43 | 6.1 | 12.1 | 3.5 | 0.6 | 18.9 | 2.6 | 4 | |
The method and origin of the coding system used in this program
The cycle and trip number, along with the species code, suffix (e.g., M. for Mollusk) and station numbers, provide necessary cross references. The cycle and trip numbers were written as 0101, 0102, 0103, etc. The first two figures in each case represent the year or cycle, and the second two, the trip number. The species codes were obtained by assigning numbers to the appropriate phylogenetic check lists, starting with 0010C, 0020C, etc. The zero between the 1 and 2 and the suffix “C” (crustaceans), was used to facilitate the insertion of species caught during the study but not included in the original check lists. Such codes would then be written as 001AC, 002AC, 003AC, etc., thus maintaining the species code numbers in phylogenetic order.
Appendix Table I
The specified format of the name card by card column.
Card color pink; one card per species per station
| Card Column | Contents |
| 1 – 70 | Printed name (i.e., Order, family, genus and species) |
| 72 – 75 | Code number of species |
| 76 | Suffix Mollusk, Fish, Crustacean, Echinoderm |
| 78 | Station number 1, 2, 3 … |
| 80 | Type of card 1 equals name |
Appendix Table II
The specified format of the data card by card column.
Card color blue; one card per species for each station and trip
| Card Column | Contents | ||
| 1 – 3 | Sample | one | Ten replicate samples maximum field size 999 individuals per sample |
| 5 – 7 | " | two | |
| 9 – 11 | " | three | |
| 13 – 15 | " | four | |
| 17 – 19 | " | five | |
| 21 – 23 | " | six | |
| 25 – 27 | " | seven | |
| 29 – 31 | " | eight | |
| 33 – 35 | " | nine | |
| 37 – 39 | " | ten | |
| 41 – 46 | Species weight in grams | ||
| 48 – 53 | Average size in mm | ||
| 55 – 57 | Minimum size in mm | ||
| 59 – 61 | Maximum size in mm | ||
| 67 – 70 | Cycle and trip number | ||
| 72 – 75 | Code number of species | ||
| 76 | Suffix Mollusk, Fish, Crustacean, Echinoderm | ||
| 78 | Station number 1, 2, 3 … | ||
| 80 | Type of card 3 equals Data | ||
Appendix Table III
The specified format of the parameter card by card column.
Card color yellow; one card per station per trip
| Card Column | Contents |
| 1 – 2 | Day of month (e.g., 12) |
| 4 – 6 | Month, (e.g., Oct.) |
| 8 – 9 | Year, (e.g., 66) |
| 11 – 14 | Salinity |
| 16 – 19 | Temperature degrees |
| 21 – 24 | Time of day (24 h clock) |
| 26 – 31 | Wind, velocity and direction during sampling |
| 33 – 38 | Vegetation weight in grams |
| 40 – 44 | Tunicate " " " |
| 46 – 50 | Sponge " " " |
| 52 – 56 | Precipitation in inches between trips |
| 58 – 62 | Evaporation in inches between trips |
| 67 – 70 | Cycle and trip number |
| 78 | Station number 1, 2, 3 … |
| 80 | Type of card 2 equals parameter |
Appendix Table IV
A listing of the fortran II program used to calculate the number of replicates (N), the sample variance, standard deviation, and the coefficient of variation. (Phase 2 Table I)
| Start of sequence | |
| PARAMI9I0508P A | |
| DIMENSION NA(10), A(10), B(10), JA(10) | |
| 1 | REWIND 4 |
| REWIND 5 | |
| READ 100, IREC | |
| 100 | FORMAT(15) |
| ITEST = 0 | |
| 2 | READ INPUT TAPE 4, 101, NA, B |
| 101 | FORMAT (13, 614, 1X, 3(13, 1X), 10A4, 53X) |
| IF(SENSE SWITCH 1) 3, 4 | |
| 3 | ILEFT = IREC - ITEST |
| PRINT 205, ITEST, ILEFT | |
| 205 | FORMAT (1H1, 15, 17HRECORDS PROCESSED, 5X, 15, 17 HRECORDS REMAINING) |
| 4 | ITEST = ITEST + 1 |
| DO 10 I = 1, 10 | |
| 10 | JA(I) = 1 |
| N = 10 | |
| NSUM = 0 | |
| DO 20 I = 1, 10 | |
| IF (NA(I)) 15, 18, 18 | |
| 15 | N = N - 1 |
| NA(I) = 0 | |
| JA(I) = 0 | |
| 18 | NSUM = NSUM + NA(I) |
| 20 | A(I) = NA(I) |
| VAR = NSUM | |
| DTF = N | |
| AVG = VAR/DTF | |
| VAR = 0.0 | |
| DO 30 I = 1, 10 | |
| IF(JA(I)) 25, 30, 25 | |
| 25 | X = A(I) - AVG |
| VAR = VAR + X*X | |
| 30 | CONTINUE |
| XN = N - 1 | |
| VAR = VAR/XN | |
| DEV = VAR**0.5 | |
| COEF = DEV/AVG | |
| DTF = 0.5*AVG*AVG | |
| REQD = 4.0*VAR/DTF | |
| IF (REQD - 1.0E4) 35, 34, 34 | |
| 34 | REQD = 999.9 |
| 35 | WRITE OUTPUT TAPE 5, 201, |
| 1 B, NSUM, AVG, VAR, DEV, COEF, DTF, REQD | |
| 201 | FORMAT(10A4, I4, 5(F6.1), F5.1, 54X) |
| IF(ITEST - IREC) 2, 40, 40 | |
| 40 | PRINT 202 |
| 202 | FORMAT(13H1JOB FINISHED) |
| PRINT 203 | |
| 203 | FORMAT(1H1) |
| END FILE 5 | |
| REWIND 5 | |
| GO TC 1 | |
| END | |
Appendix Table V
A listing of the autocoder program required to produce the printed output of Table II (summarized in Phase 3 Table I)
| Start of sequence | (Continued) | (Continued) | (Continued) | ||||||||
| JOB | LIST DATA | ABO | SBR | ABE+3 | BU | C3 | GO | SBR | GE+3 | ||
| CTL | 6611 | MLC | 179,279 | MLC | BK1,234 | SW | 1,101 | ||||
| START | B | GO | MLC | 176,CODE=5 | C3 | C | 240,ZERO | R | |||
| DATES | B | BO | BU | C4 | MLC | 5,TESTA=5 | |||||
| SW | 8,15 | W | MLC | BK2,241 | MLC | 8,TESTB=2 | |||||
| SW | 19 | W | C4 | C | 246,ZERO | R | |||||
| AGAIN | MLC | ‘0000’,)0J002 | MLC | HEAD1,236 | BU | C5 | MLC | 5,IREC=5 | |||
| MLC | ‘00000’, I TEST=5 | MLC | HEAD2,298 | MLC | BK1,246 | LCA | GK,81 | ||||
| RWD | 2 | B | BO | C5 | C | 252,ZERO | LCA | GK,181 | |||
| RWD | 5 | W | BU | C6 | SW | 41,45 | |||||
| ML1 | RWD | 3 | ABE | B | O | MLC | BK1,252 | SW | 51,57 | ||
| B | AO | ACO | SBR | ACE+3 | C6 | C | 258,ZERO | SW | 63,69 | ||
| MLC | ‘2’, UNIT=1 | MLC | HEAD3,206 | BU | CE | SW | 38,75 | ||||
| B | DO | MLC | HEAD4,214 | MLC | BK1,258 | GE | B | O | |||
| B | ABO | MLC | HEAD5,219 | CE | B | O | STOP | CC | 1 | ||
| B | ACO | MLC | HEAD6,224 | DO | SBR | DE+3 | MLC | HEAD25,275 | |||
| ML2 | MLC | ‘3’,UNIT | MLC | HEAD7,230 | MLC | UNIT,01+3 | B | BO | |||
| B | DO | MLC | HEAD8,238 | MLC | UNIT,E3+3 | W | |||||
| B | CO | MLC | HEAD9,244 | MLC | UNIT,E4+3 | W | |||||
| C | ITEST,IREC | MLC | HEAD10,250 | D1 | RT | 0,101 | MLC | HEAD26,275 | |||
| BL | ML3 | MLC | HEAD11,256 | BER | EO | B | BO | ||||
| BE | ML3 | MLC | HEAD12,262 | DE | B | O | MLC | HEAD28,275 | |||
| MLC | ‘5’,UNIT | MLC | HEAD13,267 | EO | SBR | EE+3 | B | BO | |||
| MLC | ZERO,D1+4 | MLC | HEAD14,275 | MLC | ‘50’,ATMPT=2 | CC | 1 | ||||
| MLC | ZERO,E4+4 | MLC | HEAD15,280 | E1 | MLC | ‘00’,TRY=2 | H | AGAIN | |||
| B | DO | MLC | HEAD16,284 | E2 | A | ‘1’,TRY | HEAD | DCW | ‘YEARLY ANALYSIS’ | ||
| MLC | ‘1’,01+4 | MLC | HEAD17,289 | C | TRY,ATMPT | HEAD1 | DCW | ‘PARAMETERS’ | |||
| MLC | ‘1’, E4+4 | MLC | HEAD18,295 | BL | EF | HEAD2 | DCW | ‘VARIABLES’ | |||
| C | 36,CODE | MLC | HEAD19,302 | E3 | BSP | O | HEAD3 | DCW | ‘DATE’ | ||
| BU | ML3 | MLC | HEAD20,309 | E4 | RT | 0,101 | HEAD4 | DCW | ‘SAL.’ | ||
| C | 30,TRIP=4 | MLC | HEAD21,316 | BER | E2 | HEAD5 | DCW | ‘TEMP’ | |||
| BU | ML3 | MLC | HEAD22,323 | EE | B | O | HEAD6 | DCW | ‘TIME’ | ||
| B | FO | MLC | HEAD23,328 | EF | H | HEAD7 | DCW | ‘WIND’ | |||
| B | ML4 | MLC | HEAD24,332 | BSS | E1,C | HEAD8 | DCW | ‘VG.WT.’ | |||
| ML3 | MLC | HEAD29,295 | B | BO | B | AGAIN | HEAD9 | DCW | ‘T.WT.’ | ||
| B | BO | W | FO | SBR | FE+3 | HEAD10 | DCW | ‘S.WT.’ | |||
| C | ITEST,IREC | ACE | B | O | C | 6,CHECK | HEAD11 | DCW | ‘PREC.’ | ||
| BL | ML4 | BO | SBR | BE+3 | BE | F1 | HEAD12 | DCW | ‘EVAP.’ | ||
| BE | ML4 | W | MCS | 6,269 | HEAD13 | DCW | ‘WT.’ | ||||
| BSP | 5 | CS | 332 | F1 | C | 13,CHECK | HEAD14 | DCW | ‘SIZE’ | ||
| ML4 | C | TRIP,TESTB | CS | BE | F2 | HEAD15 | DCW | ‘MIN’ | |||
| BE | ML5 | BE | B | O | MCS | 13,276 | HEAD16 | DCW | ‘MAX’ | ||
| B | ML2 | CO | SBR | CE+3 | F2 | MCS | 17,280 | HEAD17 | DCW | ‘SUM’ | |
| ML5 | C | CODE,TESTA | MLC | 162,262 | MCS | 21,284 | HEAD18 | DCW | ‘MEAN’ | ||
| BL | STOP | MLC | 170,TRIP | MLC | 44,289 | HEAD19 | DCW | ‘VAR.’ | |||
| BE | STOP | C | 201,ZERO | MLC | 50,296 | HEAD20 | DCW | ‘DEV.’ | |||
| B | ML1 | BU | C1 | MLC | 56,303 | HEAD21 | DCW | ‘C.V.’ | |||
| AO | SBR | AE+3 | MLC | BK1,201 | MLC | 62,310 | HEAD22 | DCW | ‘D.F.’ | ||
| CC | 1 | C1 | C | 228,ZERO | MLC | 68,317 | HEAD23 | DCW | ‘N’ | ||
| MLC | HEAD,275 | BU | C2 | MLC | 74,324 | HEAD24 | DCW | ‘S’ | |||
| PDATE320 | MLC | BK1,228 | MLC | 79,330 | HEAD25 | DCW | ‘LISTING COMPLETED’ | ||||
| B | BO | C2 | C | 233,ZERO | MLC | 38,332 | HEAD26 | DCW | ‘FOR ADDITIONAL LISTING’ | ||
| W | BU | C3 | B | BO | HEAD28 | DCW | ‘PUSH START’ | ||||
| W | MLC | BK1,233 | A | a-6,ITEST | HEAD29 | DCW | ‘NONE CAUGHT THIS TRIP’ | ||||
| AE | B | O | C | 234,ZERO | FE | B | O | GK | DCW | ‘ ’ | |
| BK1 | DCW | ‘ ’ | |||||||||
| ZERO | DCW | ‘0’ | |||||||||
| BK2 | DCW | ‘ ’ | |||||||||
| CHECK | DCW | ‘0000.0’ | |||||||||
| END | START | ||||||||||
Clyde, J., M. Cramer and J. Sherin, 1966 Multivariate statistical programs. Coral Gables, Florida, University of Miami, Biometrics Laboratory
Costello, T.J. and D.M. Allen, 1966 Migrations and geographic distribution of pink shrimp, Penaeus duorarum, of the Tortugas and Sanibel grounds, Florida. Fishery Bull. Fish. Wildl. Serv. U.S., 65(2):449–59
Greig-Smith, P., 1964 Quantitative plant ecology. Washington, Butterworths, 2nd.ed. 256 p.
Snedecor, W.G., 1962 Statistical methods. Iowa City, Iowa State University Press, 534 p.
Tabb, D.C. and T.M. Thomas, 1967 Environmental variation and abundance of animals in South Florida estuaries. Paper presented to the International Symposium on Coastal Lagoons, Mexico, 1967 (MS)
Tabb, D.C., D.L. Dubrow and A. Jones, 1962 Studies on the biology of pink shrimp Penaeus duorarum Burkenroad in Everglades National Park, Florida. Tech. Ser. Fla. St.Bd Conserv., (37): 31 p.