J.C.M. TRAIL, G.D.M. d'IETEREN, A. FERON, P. ITTY, O. KAKIESE, J.H.H. MAEHL, M. MULUNGO, S.M. NAGDA, R. W. PALING, M. PELO, J.M. RARIEYA and W. THORPE
Introduction
Materials and methods
Results
Discussion
References
Research on genetic improvement of trypanotolerant livestock is directed in two main areas: first, in increasing their productivity by conventional management and breeding methods; second, in determining the factors associated with their innate resistance, i.e. the control of trypanosome growth, the development of effective immune responses and resistance to anaemia. If the mechanisms underlying these factors are identified, it might be possible by immunization, by specific drug treatment or by transfection of appropriate genes to produce highly productive cattle resistant to trypanosomiasis.
Our immediate aims in the ATLN are firmly anchored in the first approach, conventional breeding for increased disease resistance. The basic initial requirements include estimation of phenotypic and genetic relationships between disease and production traits and assessing the feasibility of practical applications of genetic selection for resistance.
Two separate field experiments were utilized to provide indications of heritability levels for aspects of parasitaemia, anaemia control, immune response and animal performance.
At Mushie ranch, in Bandundu region, Zaire, 157 N'Dama progeny of four sires, 95 males and 62 females, were available in June 1987. They comprised three age groups, 21 months and 35 months for both sexes and 52 months for males only. The males were exposed to a high natural trypanosome challenge, and body weight, trypanosome aspects and PCVs were recorded weekly on eleven occasions covering a ten-week period. The females were exposed to a low natural trypanosome challenge and recorded weekly on eight occasions covering a seven-week period. The analysis approach was to estimate the heritabilities of parasitaemia aspects, anaemia control aspects and daily liveweight change.
At Kilifi Plantation! on the southeast coast of Kenya, 592 Sahiwal/Ayrshire crossbred cows were exposed to a low, natural trypanosome challenge over a five-year period and maintained under a chemotherapeutic strategy. An excellent health package for internal parasites, tick control, etc. was operated and each animal had its PCV measured four to five times per year; any animal with a PCV value of 29% or less was treated with a curative trypanocide. The trypanocidal treatment required was used as the indication of infection with trypanosomes. The analysis approach was to determine if acquired resistance existed, if it was under genetic control and if it could be used as a measure of ability to develop an effective immune response.
All traits were analysed by least-squares fixed- and mixed- model procedures (Harvey, 1977), the effects fitted being indicated in each section. Interaction effects fitted were those that preliminary analyses had indicated were significant or approached significance for any of the matching traits involved.
Mushie Ranch
The average weekly trypanosome prevalence in the male N'Dama animals over the 11 weekly recordings was 22.5%. Of the 95 males, 24.2% never had trypanosomes detected, 22.1% had trypanosomes detected in one week, 9.5% in two weeks, 15.8% in three weeks, 17.9% in four weeks and 10.5% in five or more weeks. The average weekly prevalence in the females over the eight weekly recordings was 5.1%. Of the 62 females, 85.5% never had trypanosomes detected, 4.8% had trypanosomes detected in one week, 3.2% in two weeks, 1.6% in three weeks, 3.2% in four weeks and 1.6% in five weeks.
Least-squares analyses were carried out separately for the two sexes, fitting sire and dam within sire as random effects and age group as a fixed effect. The traits evaluated over the ten-week period for males and seven-week period for females were daily liveweight change as a performance aspect; number of infections by species and total and parasitaemia score by species and total, as parasitaemia aspects; and average PCV as an anaemia control aspect.
Heritability of growth, parasitaemia aspects and PCV level
The heritability estimates for daily liveweight change, parasitaemia aspects and PCV level are indicated in Table 1. The four sires had 23, 27, 24 and 21 male progeny and 16, 14, 17 and 15 female progeny, respectively. In both high-challenge and low-challenge situations, while the heritability estimates for daily liveweight change were within the expected range, there was no indication of any of the aspects of parasitaemia being heritable. In males under a high challenge, but with a wide range in number of detected infections (from 0 to 5 or more), the heritability estimate for PCV level was 0.15, suggesting that evaluation of ability to maintain PCV levels be compared for different trypanosome prevalence levels.
Table 1. Heritability estimates for daily liveweight change, parasitaemia aspects and PCV.
|
|
Daily liveweight change |
Parasitaemia aspects (6 traits)a |
PCV |
|||
|
h2 |
s.e. |
h2 |
s.e. |
h2 |
s.e. |
|
|
Males, high tsetse challenge |
.20 |
.49 |
0 |
- |
.15. |
44 |
|
Females, low tsetse challenge |
.68 |
.63 |
0 |
- |
0 |
- |
a Total number of infections, number of T. congolense infections, number of T. vivax infections, total parasitaemia score, T. congolense parasitaemia score, T. vivax parasitaemia score.
Therefore the males were split into two groups, the 46.3% that had had trypanosomes detected in 0 or 1 week only, which represented an average trypanosome prevalence of 5% and the 53.7% that had had trypanosomes detected in two or more weeks, which represented an average trypanosome prevalence of 40%. The four sires had 12, 11, 11 and 10 progeny under low trypanosome prevalence and 11, 16, 13 and 11 under high trypanosome prevalence.
Table 2. Heritability estimates for daily liveweight change and PCV under low and high trypanosome prevalence.
|
Trypanosome prevalence groupa
|
Daily liveweight change |
PCV |
||
|
h2 |
s.e. |
h2 |
s.e. |
|
|
Low (ignoring full sibs) |
.37 |
.55 |
.01 |
.33 |
|
Low (adjusted for full sibs) |
.28 |
- |
0 |
- |
|
High (ignoring full sibs) |
.57 |
.61 |
1.10 |
.81 |
|
High (adjusted for full sibs) |
.43 |
- |
.82 |
- |
a Low trypanosome prevalence = 5%,
high = 40%
When progeny groups were subdivided, numbers were too small for dam within-sire effects to be estimated. A rerun of the analysis in Table 1 indicated that ignoring full sib effects inflated the heritability estimates by 25%. This adjustment factor was therefore used for the results in Table 2. With the heritability estimates for daily liveweight change falling within the expected range under both low and high trypanosome prevalence, the heritability of ability to maintain PCV levels was zero under no or low prevalence and 0.82 under high prevalence.
Kilifi Plantation
From 592 cows of two breed groups, 1/3 Sahiwal 2/3 Ayrshire and 2/3 Sahiwal 1/3 Ayrshire, 1308 sets of records were analyzed in a least-squares model including fixed effects of genotype, parturition number, number of previous trypanocidal treatments, year of calving, month within-year of calving and a number of interactions. Random effects included sire-within-genotype and cow-within-sire-within-genotype. The traits analyzed as indicating immune response aspects were the number of treatments required (= infections) in monthly periods after parturition. Confidence to use PCV as an indication of trypanosome infection was based both on the response to treatment and the fact that other potential anaemia-causing infections on the ranch, namely tick-borne diseases and helminthiasis, were successfully controlled by judicious dipping and strategic use of anthelmintics.
Occurrence of acquired resistance
Figure 1. Least-squares past treatment group means for number of current treatments (cumulative) for trypanosomiasis required at monthly intervals after parturition.
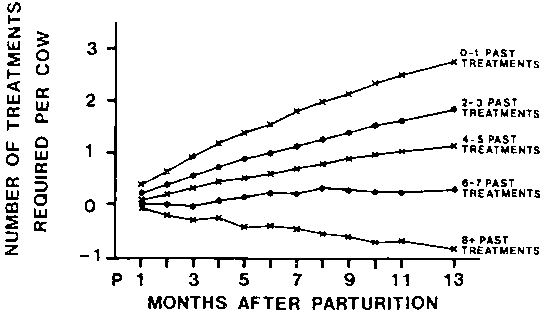
Figure 1 indicates that, when the number of trypanocidal treatments required was used as an indication of infection, resistance was being acquired. The evidence was that the more treatments an animal had had previously, the less treatments it was liable to require in future. For example any animal with 0 to 1 past treatments would need 3 treatments during the next postpartum period, whereas, any animal with 6 or more past treatments would not require any treatment.
Genotype differences in acquired resistance
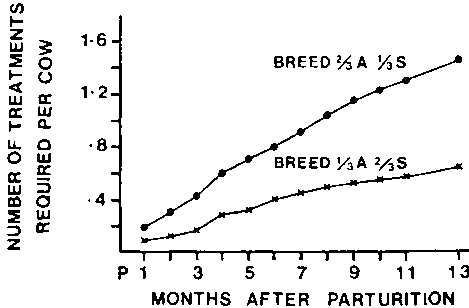
There were significant differences between genotypes in the number of treatments required (Figure 2). The 2/3 Sahiwal required less than half the treatments of the 1/3 Sahiwal over the complete calving interval; by the end of this period the 2/3 Sahiwal needed 0.53 treatments while the 1/3 Sahiwal required 1.45. A significant interaction between genotypes and previous treatments showed that when very few previous treatments (0 to 1) had been received, both genotypes required similar numbers of current treatments. However, as the number of previous treatments increased, the 1/3 Sahiwal required progressively more current treatments than the 2/3 Sahiwal, suggesting that the 2/3 Sahiwal acquired resistance better than the 1/3 Sahiwal.
Figure 2. Least-squares breed group means for number of current treatments (cumulative) for trypanosomiasis required at monthly intervals after parturition.
Heritability of acquired resistance
The 739 records of the 335 cows of the 2/3 Sahiwal group that appeared more able to acquire resistance were divided into two groups, those with a low (0 to 1) number of previous treatments (406 records) and those with a high (2 to 10) number (333 records). The two groups were analysed separately in a least-squares model including fixed effects of parturition number and year. Random effects included sire (16 in number) and cow within sire. The traits analyzed were the number of treatments required (= infections) in four periods after parturition (3, 6, 9 months and calving interval). The heritability estimates are shown in Table 3.
Table 3. Heritability estimates for number of treatments required (= infections) in four periods after parturition.
|
Period after parturition |
Number of previous treatments (= infections) |
|
|
Low (0 or 1) |
High (2 - 10) |
|
|
3 months |
0 |
0.01 + 0.11 |
|
6 months |
0 |
0.09 + 0.14 |
|
9 months |
0 |
0.26 + 0.20 |
|
Complete calving interval |
0 |
0.23 + 0.19 |
When there had been zero or 1 previous treatment, the heritability of the number of treatments required for all four periods after parturition was zero. In contrast, when there had been a high number of previous treatments, the heritability increased with the time period involved.
From the studies at Mushie Ranch and Kilifi Plantation, key aspects designed to provide indications of heritability levels of parasitaemia, anaemia control and immune response, are indicated in Table 4.
Table 4. Indications of heritability levels for criteria of trypanotolerance at Mushie Ranch and Kilifi Plantation.
|
Criteria |
Heritability | |
|
Parasitaemia aspects | ||
|
|
Number of infections per unit time |
0 |
|
|
Infection score per unit time |
0 |
|
|
T. congolense number and score |
0 |
|
|
T. vivax number and score |
0 |
|
Anaemia control aspects | ||
|
|
PCV levels overall |
0.15 |
|
|
PCV levels when no, or low, trypanosome prevalence |
0 |
|
|
PCV levels when high trypanosome prevalence |
0.82 |
|
Immune response aspects | ||
|
|
Number of infections in 9 months when few previous infections |
0 |
|
|
Number of infections in 9 months when many previous infections |
0.26 |
These first results suggest that these particular parasitaemia aspects are unlikely to have sufficiently high heritabilities for practical selection approaches; that ability to maintain PCV levels under high natural challenge could well be the basis of a practical selection approach for anaemia control; and that in certain circumstances it might be possible to select for ability to acquire resistance to trypanosomiasis.
Considerable efforts continue to be put into building up paternal half-sib groups in situations in the ATOM, using both single-sire matings and blood-grouping techniques. The goal is to significantly increase information on heritability estimates and relevant genetic correlations.
Harvey, W.R. 1977. User's Guide for Least-Squares and Maximum Likelihood Computer Program. Columbus: Ohio State University.