A. J. TEALE and S. J. KEMP
Introduction
MHC typing of cattle
MHC studies in N'Dama cattle
Materials and methods
Results
Conclusion
References
The MHC comprises a series of genes encoding glycoproteins which are expressed on the surface of cells. The genes are physically associated on a single chromosome and tend to be inherited as a group. In most mammalian species there is a considerable number of such genes, although not all have detectable products at the cell surface.
The genes and their products can be placed into two distinct groups. Class I gene products are expressed on the surface of all nucleated cells, whereas class II gene product expression is largely limited to the surface of some cells of the immune system with rather specialized functions. Class I and class II gene products can also be distinguished on the basis of their biochemical nature.
An important feature of the MHC is its extreme polymorphism; i.e., the degree to which the individual genes and their products vary between unrelated individuals of a species. The number of loci and alleles at the different loci in man for instance, is such that on theoretical grounds no two unrelated people will have an identical set of MHC-encoded glycoproteins on the surface of their cells. Moreover, expression of MHC genes is codominant, and so each individual expresses one set, or haplotype, inherited from the mother and one haplotype inherited from the father.
The function of MHC glycoproteins is to bind and present foreign antigens (proteins of non-self origin to which an immune response is mounted) to cells of the immune system so that an appropriate immune response can be generated. Parts of proteins (peptides), of viruses, bacteria and protozoa, for instance, physically associate with MHC glycoproteins and then become recognizable by the immune system cells. Once these cells encounter the foreign peptide/MHC glycoprotein complex antigen, they become functionally activated and proliferate and, through various mechanisms eliminate the invading pathogen. However, a given MHC glycoprotein will not be able to associate with, and so present, all of the potentially immunogenic peptides which may be generated within an animal during its lifetime. Consequently, the MHC profile of an individual can have a major effect on the immune response repertoire. Further, the MHC profile can result in the failure of the immune response as a consequence of mimicry of a particular peptide/MHC molecule combination by a second MHC molecule, per se, within the same animal. As animals are "tolerant" (a state in which immune responses are not mounted) of their own proteins, which include their MHC molecules, in this circumstance there can be no immune recognition of the peptide concerned.
In view of the influential nature of the MHC in generation of immune responses, it might be expected that disease susceptibility may be associated with the MHC profiles, or "types" of individuals within a species; that is, that individuals with certain MHC types exist which are either more or less likely than the average of the species to suffer a given disease. In man, for instance, there is now an impressive list of diseases which show significant associations with MHC type (Tiwari and Terasaki 1985). Although many such associations involve non-fatal chronic diseases, associations with important infectious diseases have been reported, including malaria (Piazza et al., 1976), typhoid and yellow fever (De Vries et al., 1979), poliomyelitis (Van Eden et al., 1983) and leprosy (Van Eden et al., 1982).
Clearly, in order to make strong associations in the first instance, an ability to characterize the MHC in all its diversity is a prerequisite. It is now possible to define polymorphism in both the class I and class II regions in cattle. In the case of class I products, these are universally characterized in a standard microlymphocytotoxicity test which is based on the reactivity of the cells of an animal to be typed with panels of antisera and monoclonal antibodies. When a class I gene product is recognized by an antibody in a panel, the cell carrying the MHC molecule is killed and the cytotoxicity measured. The overall pattern of reactivity of the cells of an individual animal with the panel of typing reagents gives the class I phenotype of the animal concerned. Through international collaborative efforts the definition of the bovine MHC has advanced to the point where more than 20 bovine class I gene products are now recognized by all laboratories working in this area. Over the past few years, in collaboration with scientists at the AFRC Institute of Animal Physiology and Genetics Research, Edinburgh, we have been working at ILRAD to improve the MHC typing of African cattle breeds in particular. Now, in addition to those MHC types recognized internationally, we are able to define MHC gene products which are apparently characteristic of African breeds (Kemp et al., 1988).
We and others are also able to characterise class I and II gene products with other techniques which include biochemical and cellular methods. The class II region is proving definable at the level of the DNA of the genes themselves, using the technique of restriction fragment length polymorphism (RFLP) analysis. All of these non-serological approaches are somewhat more tedious and technically demanding than the standard microlymphocytotoxicity assay, which is therefore likely to remain the most widely applied for some considerable time.
Although several class I loci are defined in man and laboratory animals, it is still not clear whether all of the class I types now recognized in cattle are encoded by a single locus or by multiple loci. However, this ignorance of the complexity of bovine MHC haplotypes need not necessarily delay searches for associations between MHC type and disease resistance/susceptibility. This is because, given that there is a considerable degree of linkage over the length of the MHC, identification of any of the products of an haplotype and its differentiation from the products of other haplotypes in a study population, is all that is required to follow MHC inheritance.
The trypanotolerant trait of some West African races of Bos taurus cattle is well documented but the genetic control is unclear. We have begun a study of the MHC of N'Dama in The Gambia which involves collecting sufficient typing data to enable a valid comparison of the MHC profiles of these cattle with those of unrelated "susceptible" breeds. In addition, we have attempted to identify extremes within the N'Dama population, with respect to trypanotolerance, to enable a within-breed comparison. The overall objective is to reveal haplotypes which may be associated with trypanotolerance for further study in laboratory challenge experiments prior to detailed analysis of the individual genes carried on such haplotypes.
MHC typing
This was performed essentially as reported previously (Teale et al. 1983) with modifications required by an automated reading system (Kemp and Teale, 1987).
Cattle populations
All of the N'Dama animals are under study at the ITC in The Gambia. The population sample for typing is made up of two groups:
A. Cattle in the Livestock Development Project (LDP) herds at Kerr-Serigne and Keneba. These animals may be considered to be representative of the Gambian population of N'Damas as they were acquired from a number of different herds in various parts of the country. They will form the nucleus of the LDP breeding programme. This group consists of approximately 270 cows, calves and bulls.B. Approximately 50 present and future breeding bulls in 4 village locations. The bulls can be considered to be representative of the 2500 or so cattle being monitored for various production/performance characteristics under village management systems. Disease data are also being collected in this population. The bulls were specifically chosen because they may be expected to have a major impact on the future MHC structures of the village herds.
Sample preparation
Peripheral blood mononuclear cells (PBM) for typing were either separated from whole blood samples and placed into liquid nitrogen at the ITC using a standard method of cryopreservation, or aliquots of whole blood samples were cryopreserved as previously described (Kemp and Teale, 1984) for subsequent preparation of PBM at ILRAD.
To date, the cells of more than 300 N'Damas have been tested with our panel of 180 class I typing reagents. Antigen frequencies have been compared with those in other populations of cattle. The frequencies for a number of them and including all those which show significant frequency differences between breeds, are shown in Figure 1. The frequency of the null allele (i.e., genes for which no product is detected) in the European, Boran and N'Dama populations is 0.58, 0.44 and 0.54, respectively.
Figure 1. Frequencies of BoLA class I antigens in three races of cattle.
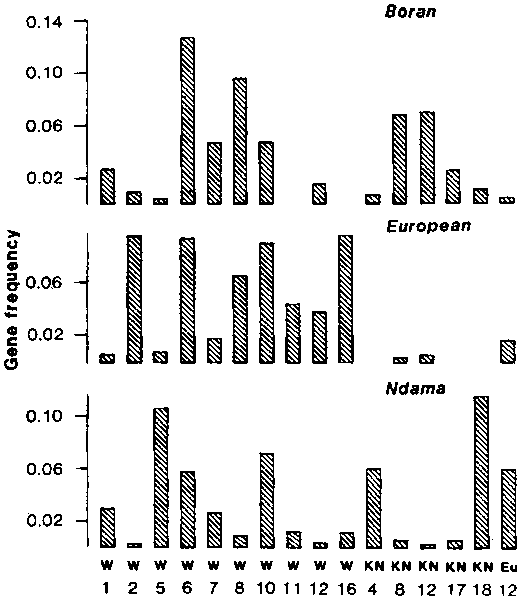
With respect to features of the MHC profiles which seem characteristic of the N'Dama population, the most striking involves the KN18 antigen which is present in 21% of the N'Dama cattle but in only 1.5% of the Borans and in only one of the European animals. This is a very well characterized antigen and such a difference in its frequency between N'Dama and other cattle breeds is worthy of further investigation.
Another well-defined specificity is w10 which has been shown to have two subgroups, KN3 and KN104 (Kemp et al., 1988). These subgroups have never been observed in European cattle but are common in African Boran and Zebu breeds. Most N'Dama w10 animals were found to have both of these subgroups and so could be considered to possess typical "African" w10 antigens. However, individuals with w10 but lacking both subgroups were found among the N'Dama. This is typical of the w10 antigen in European cattle and is extremely rare in Bos indicus animals.
Like Boran cattle, the N'Dama were found to have a low frequency of w2, an antigen which is common in European breeds.
Other striking features of the N'Dama BoLA profile are the relatively high frequency of KN4 and the low frequency of KN8 and w8. The N'Dama also appeared to have a very high frequency of w3 and w5. However, we believe that the w3 and w5 antigens are not being reliably detected in the N'Dama. Therefore, although the differences in the frequency of the antigens detected is real and possibly significant, the w3 and w5 nomenclature should not be taken to mean that the differences are in fact in frequencies of internationally-agreed antigens.
Recently we have reported a detailed comparative analysis of the MHC gene products of European taurine and African zebu types (Kemp et al., 1988). It is now apparent that there are clear differences in the BoLA profiles of the Gambian N'Dama by comparison with the European and East African animals and each of the major breed groups described therefore has some unique features in its BoLA profile. Thus the high frequency of KN18 is a particular feature of the N'Dama population whereas high frequencies of KN8 and KN12 are more typical of the East African Zebus. In some respects however, there are indications of similarities between the N'Dama population and the Zebu population on the one hand and between the N'Damas and the European cattle, on the other. This is illustrated by the occurrence of the KN104 gene in both the N'Dama and Zebu populations but not in the European breeds and by the occurrence of a particular w10 subtype in the N'Dama and European cattle which is not seen in the Zebus. In some cases there are no significant differences in frequencies within the three groups.
Differences where they are seen between distinct populations within a species are not unexpected and have been well documented in human races, for example (Baur and Danilovs, 1980). Whether the differences seen in the cattle populations are the result of selection for advantageous types in the different environments or whether they are the result of random drift is not clear at the present time.
Nevertheless, these results are of value in that they make it possible to select haplotypes for further studies of the involvement of the MHC in trypanotolerance, on a logical rather than on a random basis. The segregation of selected haplotypes would be followed in animals in which response to parasite challenge is monitored. Bulls carrying typically N'Dama haplotypes (e.g., KN18) and haplotypes with low frequency in the population as a whole would be used to produce half-sibling families in matings with cows chosen on the same basis. In this way calves with various combinations of "N'Dama" and "non-N'Dama" haplotypes would be made available for field and laboratory challenge studies.
Bulls selected for heterozygosity as described would also be obvious choices for matings with cows of non-trypanotolerant type, producing F1 and ultimately F2 animals which could be used in challenge experiments to further investigate whether or not the bovine MHC has an influence on the trypanotolerance trait.
Acknowledgements
We would like to thank our colleagues and staff at the ITC for their help in planning, collection and transportation of the Gambian samples and for the hospitality shown to us during our visits to the Gambia. In particular, we would like to mention Ron Dwinger, Alistair Grieve, Phillipe Jeannin, Kwaku Agyemang, Derek Clifford and the Director of the ITC, Professor Ian McIntyre. This is ILRAD publication number 595.
Baur M.P. and J.A. Danilovs. 1980. Population analysis of HLA-A, B. C, and/or other genetic markers. In: Histocompatibility Testing. P.I. Terasaki, ed. UCLA Tissue Typing Laboratory, Los Angeles, pp. 955-993.
De Vries R.R.P., P. Meera Khan, L.F. Bernini, E. van Loghem and J.J. van Rood. 1979. Genetic control of survival in epidemics. J. Immunogen. 6: 271-287.
Kemp S.J. and A.J. Teale. 1987. An assay for the fully automated reading of the lymphocytotoxicity test and its application to the detection of Bovine Class I and Class II MHC antigens. Animal Genetics 18: (suppl. 1), 15-16.
Kemp, S.J. and A.J. Teale. 1984. Cryopreservation of lymphocytes in whole cattle blood: A method suited to the field collection of large numbers of samples. Animal Blood Groups and Biochemical Genetics 15: 219-222.
Kemp S.J., R.L. Spooner and A.J. Teale. 1988. A comparative study of major histocompatibility complex antigens in East African and European cattle breeds. Animal Genetics. 19: 17-29.
Piazza A., M.C. Belvedere, D. Bernoco D, C. Conighi, L. Contu, E.S. Curtoni, P.L. Mattiuz, W. Mayr, P. Richiardi, G. Scudeller and R. Ceppellini. 1973. In: Histocompatibility Testing 1972. J. Dausset and J. Colombani, eds., Copenhagen: Munksgaard, pp. 73-84.
Teale, A.J., S.J. Kemp, F. Young and R.L. Spooner. 1983. Selection by major histocompatibility lyre (BoLA), of lymphoid cells derived from a bovine chimard and transformed by Theileria parasites. Parasite Immunology 5: 329-336.
Tiwari J.L. and P.I. Terasaki. 1985. HLA and Disease Associations. New York: Springer-Verlag.
Van Eden W., R.R.P. De Vries, J. D'Amaro, G.M.Th. Schreuder, D.L. Leiker and J.J. van Rood. 1982. HLA-DR associated genetic control of the type of leprosy in a population from Surinam. Human Immunology 4: 343-350.
Van Eden W., G.G. Persijn., H. Bijkerk, R.R.P. de Vries, R.K.B. Schuurman and J.J. van Rood. 1983. Differential resistance to paralytic poliomyelitis controlled by histocompatibility leukocyte antigens. J. Infect. Dis. 147: 422-426.